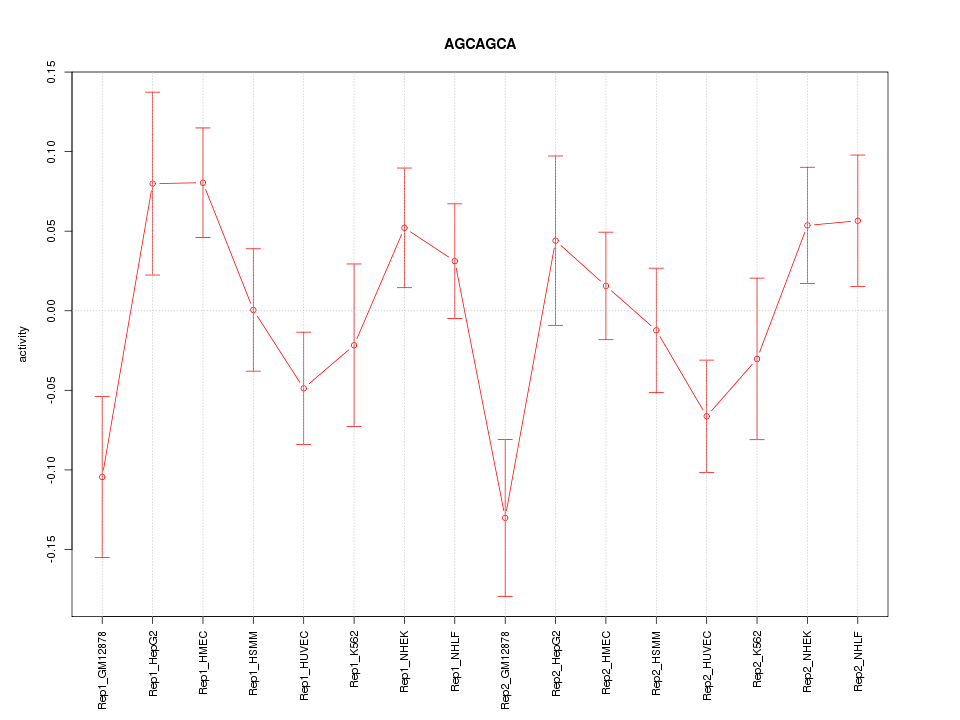
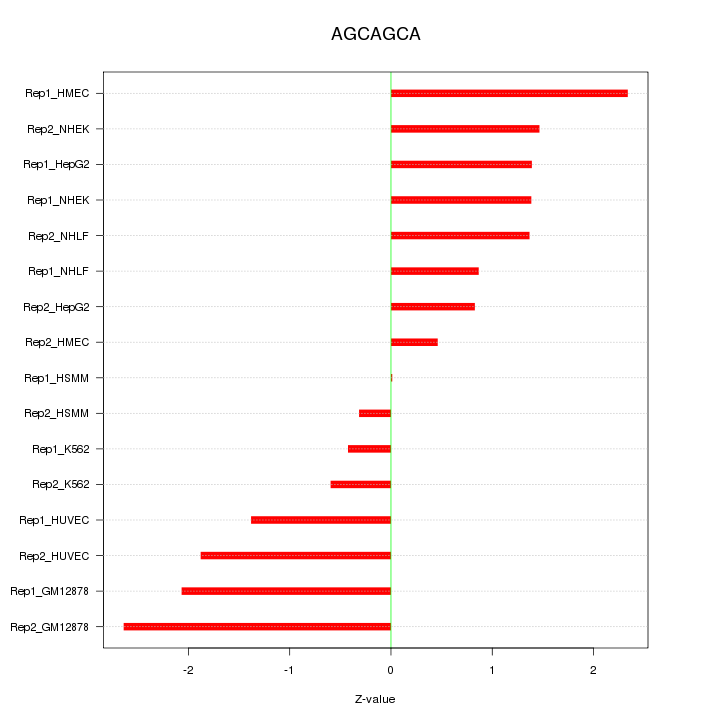
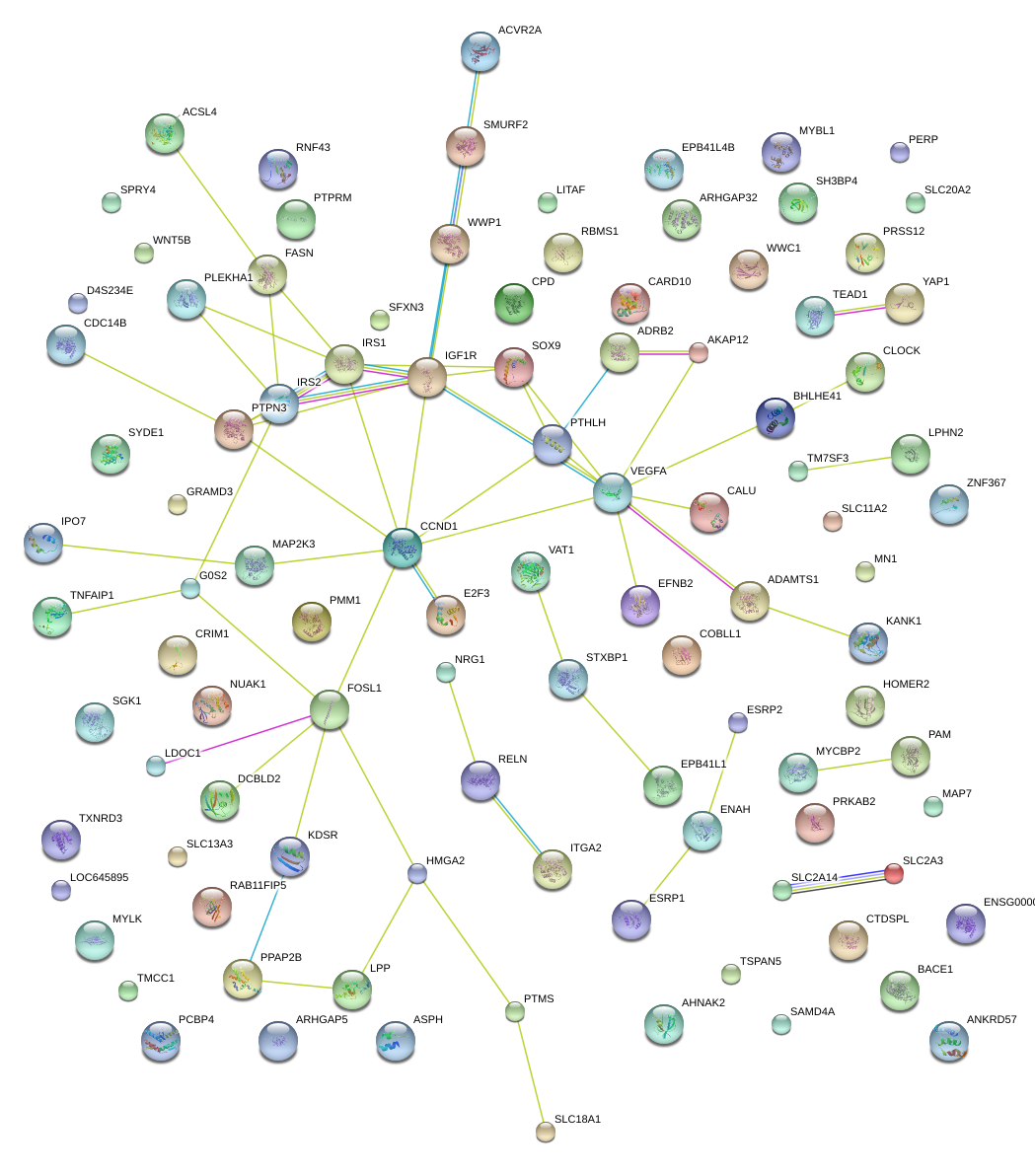
|
chr2_-_227663454
|
3.076
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr11_+_69455837
|
2.924
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr9_+_706744
|
2.612
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr9_+_504661
|
2.546
|
NM_015158
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr2_-_161350304
|
2.507
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr1_-_225840660
|
2.003
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr2_+_110371887
|
2.001
|
NM_023016
|
ANKRD57
|
ankyrin repeat domain 57
|
|
chr10_+_124145584
|
1.952
|
NM_001195608
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr6_-_134498973
|
1.937
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr3_-_98620452
|
1.897
|
NM_080927
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr6_-_134639195
|
1.891
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr3_-_123603144
|
1.861
|
NM_053025
NM_053026
NM_053027
NM_053028
|
MYLK
|
myosin light chain kinase
|
|
chr3_-_123339423
|
1.826
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr6_-_134496006
|
1.815
|
NM_005627
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr3_+_37903643
|
1.744
|
NM_001008392
NM_005808
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chr3_+_187871473
|
1.692
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr10_+_124151818
|
1.689
|
NM_021622
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr10_+_124134093
|
1.666
|
NM_001001974
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr9_-_99329092
|
1.655
|
NM_001077181
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr6_-_134497069
|
1.636
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr11_+_101980736
|
1.588
|
NM_001130145
NM_001195044
NM_006106
|
YAP1
|
Yes-associated protein 1
|
|
chr9_-_112213663
|
1.572
|
NM_001145369
NM_001145370
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr3_+_187943192
|
1.554
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr9_+_130374466
|
1.543
|
NM_001032221
NM_003165
|
STXBP1
|
syntaxin binding protein 1
|
|
chr3_+_187930720
|
1.522
|
NM_005578
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr21_-_28217692
|
1.494
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr1_-_57045235
|
1.493
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr15_+_99192696
|
1.487
|
NM_000875
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr9_-_112191105
|
1.486
|
NM_001145371
NM_001145372
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr11_+_101983170
|
1.473
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr11_+_12695851
|
1.447
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr1_+_209848659
|
1.409
|
NM_015714
|
G0S2
|
G0/G1switch 2
|
|
chr6_+_151561094
|
1.374
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr8_+_95653340
|
1.351
|
NM_001034915
NM_001122825
NM_001122826
NM_001122827
NM_017697
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr9_-_112083005
|
1.341
|
NM_018424
NM_019114
|
EPB41L4B
|
erythrocyte membrane protein band 4.1 like 4B
|
|
chr11_-_65667860
|
1.289
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr14_-_105444693
|
1.263
|
NM_138420
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr16_-_68269948
|
1.240
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr9_-_99382073
|
1.237
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr8_-_67525426
|
1.232
|
NM_001080416
NM_001144755
|
MYBL1
LOC645895
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 1
uncharacterized LOC645895
|
|
chr2_+_36583369
|
1.190
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr14_+_32546445
|
1.186
|
NM_001030055
NM_001173
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr5_+_125758818
|
1.180
|
NM_001146320
NM_001146321
NM_023927
|
GRAMD3
|
GRAM domain containing 3
|
|
chr6_+_151646634
|
1.178
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr4_-_99579540
|
1.169
|
NM_005723
|
TSPAN5
|
tetraspanin 5
|
|
chr5_+_125695787
|
1.103
|
NM_001146319
|
GRAMD3
|
GRAM domain containing 3
|
|
chr17_+_28705922
|
1.096
|
NM_001304
|
CPD
|
carboxypeptidase D
|
|
chr3_-_129599220
|
1.094
|
NM_001017395
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr12_+_66218141
|
1.077
|
NM_003483
NM_003484
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr5_+_167718485
|
1.072
|
NM_001161661
NM_001161662
NM_015238
|
WWC1
|
WW and C2 domain containing 1
|
|
chr8_+_32504250
|
1.064
|
NM_013959
|
NRG1
|
neuregulin 1
|
|
chr8_+_31497267
|
1.058
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr17_-_62658311
|
1.027
|
NM_022739
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2
|
|
chr17_+_28707495
|
1.021
|
NM_001199775
|
CPD
|
carboxypeptidase D
|
|
chr8_-_42397048
|
1.018
|
NM_006749
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2
|
|
chr9_-_112260529
|
1.016
|
NM_001145368
NM_002829
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr5_-_141704575
|
1.011
|
NM_001127496
NM_030964
|
SPRY4
|
sprouty homolog 4 (Drosophila)
|
|
chr5_+_125800786
|
1.002
|
NM_001146322
|
GRAMD3
|
GRAM domain containing 3
|
|
chr13_-_110438896
|
1.000
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr1_+_82266081
|
0.988
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr17_-_41174431
|
0.978
|
NM_006373
|
VAT1
|
vesicle amine transport protein 1 homolog (T. californica)
|
|
chr5_+_52284903
|
0.956
|
NM_002203
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor)
|
|
chr10_+_102790984
|
0.951
|
NM_030971
|
SFXN3
|
sideroflexin 3
|
|
chr18_+_7567313
|
0.934
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chrX_-_140271293
|
0.933
|
NM_012317
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr17_+_70117154
|
0.916
|
NM_000346
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr17_-_80056104
|
0.910
|
NM_004104
|
FASN
|
fatty acid synthase
|
|
chr2_+_235860616
|
0.909
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr13_-_107187312
|
0.885
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr12_-_51403028
|
0.883
|
NM_001174130
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr11_+_9406123
|
0.879
|
NM_006391
|
IPO7
|
importin 7
|
|
chr5_+_102201526
|
0.876
|
NM_000919
NM_001177306
NM_138766
NM_138821
NM_138822
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr22_-_41985810
|
0.871
|
NM_002676
|
PMM1
|
phosphomannomutase 1
|
|
chr1_-_146644045
|
0.870
|
NM_005399
|
PRKAB2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit
|
|
chr11_-_128894008
|
0.862
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr3_-_126373957
|
0.845
|
NM_001173513
NM_052883
|
TXNRD3
|
thioredoxin reductase 3
|
|
chr12_+_1726221
|
0.836
|
NM_030775
|
WNT5B
|
wingless-type MMTV integration site family, member 5B
|
|
chr17_-_56494930
|
0.831
|
NM_017763
|
RNF43
|
ring finger protein 43
|
|
chr19_+_15218141
|
0.820
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr7_-_103629962
|
0.817
|
NM_005045
NM_173054
|
RELN
|
reelin
|
|
chr20_+_34700257
|
0.813
|
NM_177996
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr22_-_37915209
|
0.798
|
NM_014550
|
CARD10
|
caspase recruitment domain family, member 10
|
|
chr8_+_87354958
|
0.798
|
NM_007013
|
WWP1
|
WW domain containing E3 ubiquitin protein ligase 1
|
|
chr22_-_36236421
|
0.795
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr12_-_51419923
|
0.794
|
NM_001174129
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr18_-_61034350
|
0.794
|
NM_002035
|
KDSR
|
3-ketodihydrosphingosine reductase
|
|
chr6_-_138428477
|
0.774
|
NM_022121
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr5_+_148206155
|
0.771
|
NM_000024
|
ADRB2
|
adrenergic, beta-2-, receptor, surface
|
|
chr7_+_128379345
|
0.765
|
NM_001130674
NM_001199671
NM_001199672
NM_001199673
NM_001199674
NM_001219
|
CALU
|
calumenin
|
|
chr12_-_106533810
|
0.756
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr2_+_148602569
|
0.755
|
NM_001616
|
ACVR2A
|
activin A receptor, type IIA
|
|
chr12_-_27167332
|
0.750
|
NM_016551
|
TM7SF3
|
transmembrane 7 superfamily member 3
|
|
chr11_-_117166247
|
0.739
|
NM_001207048
NM_001207049
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr8_-_62627167
|
0.738
|
NM_001164754
NM_001164755
NM_001164756
NM_004318
NM_032466
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr3_-_129612371
|
0.734
|
NM_001128224
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chrX_-_108976509
|
0.730
|
NM_004458
NM_022977
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr4_+_4387982
|
0.729
|
NM_001040101
NM_014392
|
D4S234E
|
DNA segment on chromosome 4 (unique) 234 expressed sequence
|
|
chr4_-_56412968
|
0.723
|
NM_004898
|
CLOCK
|
clock homolog (mouse)
|
|
chr3_-_51996855
|
0.723
|
NM_033010
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr12_-_26277807
|
0.715
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr12_+_1738411
|
0.705
|
NM_032642
|
WNT5B
|
wingless-type MMTV integration site family, member 5B
|
|
chr6_-_136847743
|
0.705
|
NM_001198614
NM_001198618
NM_001198619
|
MAP7
|
microtubule-associated protein 7
|
|
chr4_-_119273874
|
0.704
|
NM_003619
|
PRSS12
|
protease, serine, 12 (neurotrypsin, motopsin)
|
|
chr17_+_26662547
|
0.703
|
NM_021137
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial)
|
|
chr2_-_73340073
|
0.703
|
NM_015470
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr20_-_45313122
|
0.703
|
NM_001011554
NM_001193340
|
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3
|
|
chr14_+_55034329
|
0.700
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr12_-_8088629
|
0.694
|
NM_006931
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3
|
|
chr6_+_20402040
|
0.688
|
NM_001949
|
E2F3
|
E2F transcription factor 3
|
|
chr12_-_28122884
|
0.681
|
NM_198964
NM_198966
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr22_-_28197469
|
0.679
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr2_-_165697927
|
0.665
|
NM_014900
|
COBLL1
|
COBL-like 1
|
|
chr6_+_43737937
|
0.651
|
NM_001025366
NM_001025367
NM_001025368
NM_001025369
NM_001025370
NM_001033756
NM_001171622
NM_001171623
NM_001171624
NM_001171625
NM_001171626
NM_001171627
NM_001171628
NM_001171629
NM_001171630
NM_001204384
NM_001204385
NM_003376
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr16_-_11680756
|
0.650
|
NM_001136472
NM_001136473
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr17_+_21191347
|
0.648
|
NM_002756
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr9_+_132962871
|
0.642
|
NM_001128826
|
NCS1
|
neuronal calcium sensor 1
|
|
chr12_-_28124915
|
0.638
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr14_-_55878537
|
0.633
|
NM_014924
|
ATG14
|
ATG14 autophagy related 14 homolog (S. cerevisiae)
|
|
chr22_-_36424397
|
0.632
|
NM_001082578
NM_001082579
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr6_+_107811272
|
0.632
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr11_-_129062092
|
0.631
|
NM_001142685
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr20_-_45280059
|
0.631
|
NM_001193339
NM_001193342
NM_022829
|
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3
|
|
chr17_-_2304217
|
0.626
|
NM_020310
|
MNT
|
MAX binding protein
|
|
chr19_-_39443326
|
0.623
|
NM_148169
|
FBXO17
|
F-box protein 17
|
|
chr3_+_57994087
|
0.619
|
NM_001164317
NM_001164318
NM_001164319
NM_001457
|
FLNB
|
filamin B, beta
|
|
chr16_+_90015827
|
0.613
|
NM_001242819
|
DEF8
|
differentially expressed in FDCP 8 homolog (mouse)
|
|
chr8_-_122653493
|
0.603
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr17_+_41322413
|
0.600
|
NM_031862
NM_031858
|
NBR1
|
neighbor of BRCA1 gene 1
|
|
chr16_+_69220940
|
0.599
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr9_+_36036900
|
0.598
|
NM_021111
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs
|
|
chr16_-_11680146
|
0.594
|
NM_004862
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr16_-_30798206
|
0.593
|
NM_001080417
|
ZNF629
|
zinc finger protein 629
|
|
chr20_+_3776385
|
0.591
|
NM_004358
NM_021872
NM_021873
|
CDC25B
|
cell division cycle 25 homolog B (S. pombe)
|
|
chr12_-_8025419
|
0.588
|
NM_153449
|
SLC2A14
|
solute carrier family 2 (facilitated glucose transporter), member 14
|
|
chrX_-_20284708
|
0.588
|
NM_004586
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr12_+_57984914
|
0.587
|
NM_001146258
NM_001146259
NM_001146260
NM_024779
|
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma
|
|
chr6_-_136847078
|
0.582
|
NM_001198608
NM_001198609
NM_001198611
|
MAP7
|
microtubule-associated protein 7
|
|
chr3_-_145968958
|
0.580
|
NM_001128306
NM_001177304
NM_020353
|
PLSCR4
|
phospholipid scramblase 4
|
|
chr19_-_39466290
|
0.579
|
NM_024907
|
FBXO17
|
F-box protein 17
|
|
chr14_-_53417808
|
0.570
|
NM_001134999
NM_001135000
NM_006832
|
FERMT2
|
fermitin family member 2
|
|
chr2_-_55277698
|
0.570
|
NM_020532
NM_153828
NM_207520
|
RTN4
|
reticulon 4
|
|
chr17_-_1389013
|
0.570
|
NM_001080950
|
MYO1C
|
myosin IC
|
|
chr16_+_14726667
|
0.563
|
NM_016561
|
BFAR
|
bifunctional apoptosis regulator
|
|
chr6_-_136871956
|
0.563
|
NM_001198616
NM_001198617
NM_003980
|
MAP7
|
microtubule-associated protein 7
|
|
chr11_-_117186918
|
0.563
|
NM_012104
NM_138971
NM_138972
NM_138973
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr2_-_122042767
|
0.562
|
NM_014553
|
TFCP2L1
|
transcription factor CP2-like 1
|
|
chr7_+_129007942
|
0.560
|
NM_001130723
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr19_-_51522953
|
0.558
|
NM_145888
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr2_-_97405799
|
0.556
|
NM_001142292
NM_030805
|
LMAN2L
|
lectin, mannose-binding 2-like
|
|
chr11_-_115375065
|
0.554
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chrX_+_128674181
|
0.550
|
NM_000276
NM_001587
|
OCRL
|
oculocerebrorenal syndrome of Lowe
|
|
chr4_+_154074269
|
0.550
|
NM_001130067
|
TRIM2
|
tripartite motif containing 2
|
|
chr17_-_1395950
|
0.549
|
NM_001080779
|
MYO1C
|
myosin IC
|
|
chr1_+_182992329
|
0.544
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr14_+_24899139
|
0.543
|
NM_015299
|
KHNYN
|
KH and NYN domain containing
|
|
chr8_-_62602285
|
0.541
|
NM_001164750
NM_001164751
NM_001164752
NM_001164753
NM_020164
NM_032467
NM_032468
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr17_-_35656691
|
0.534
|
NM_198837
NM_198838
|
ACACA
|
acetyl-CoA carboxylase alpha
|
|
chr1_-_120612301
|
0.530
|
NM_001200001
NM_024408
|
NOTCH2
|
notch 2
|
|
chr4_+_154125589
|
0.528
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr2_-_220094354
|
0.528
|
NM_001077198
NM_024085
|
ATG9A
|
ATG9 autophagy related 9 homolog A (S. cerevisiae)
|
|
chrX_+_49687212
|
0.517
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr9_-_27529434
|
0.515
|
NM_024761
|
MOB3B
|
MOB kinase activator 3B
|
|
chr6_+_143999058
|
0.508
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr12_+_54332575
|
0.506
|
NM_017410
|
HOXC13
|
homeobox C13
|
|
chr17_-_35766762
|
0.503
|
NM_198834
NM_198839
|
ACACA
|
acetyl-CoA carboxylase alpha
|
|
chr7_+_128864740
|
0.500
|
NM_001130720
NM_015328
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr3_+_171757413
|
0.492
|
NM_022763
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr7_-_123388711
|
0.491
|
NM_003941
|
WASL
|
Wiskott-Aldrich syndrome-like
|
|
chr17_-_35716024
|
0.490
|
NM_198836
|
ACACA
|
acetyl-CoA carboxylase alpha
|
|
chr18_+_67956136
|
0.482
|
NM_004232
|
SOCS6
|
suppressor of cytokine signaling 6
|
|
chr2_-_230135954
|
0.477
|
NM_001100818
NM_017933
|
PID1
|
phosphotyrosine interaction domain containing 1
|
|
chr12_-_51422012
|
0.475
|
NM_001174125
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr2_+_198381021
|
0.475
|
NM_001204094
|
MOB4
|
MOB family member 4, phocein
|
|
chr17_+_46125675
|
0.474
|
NM_003204
|
NFE2L1
|
nuclear factor (erythroid-derived 2)-like 1
|
|
chr10_+_101419169
|
0.470
|
NM_020354
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr18_-_30050394
|
0.462
|
NM_001242409
NM_022751
|
FAM59A
|
family with sequence similarity 59, member A
|
|
chr5_+_150827138
|
0.461
|
NM_078483
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr1_+_2160133
|
0.459
|
NM_003036
|
SKI
|
v-ski sarcoma viral oncogene homolog (avian)
|
|
chr6_+_143929316
|
0.459
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr17_-_1394849
|
0.453
|
NM_033375
|
MYO1C
|
myosin IC
|
|
chr5_-_95297597
|
0.453
|
NM_012081
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr11_-_35441099
|
0.452
|
NM_004171
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr4_+_72204769
|
0.448
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr19_-_5978225
|
0.448
|
NM_003624
NM_007320
NM_007322
|
RANBP3
|
RAN binding protein 3
|
|
chr6_-_136788012
|
0.444
|
NM_001198615
|
MAP7
|
microtubule-associated protein 7
|
|
chr11_+_86748870
|
0.444
|
NM_001168724
NM_022918
|
TMEM135
|
transmembrane protein 135
|
|
chr6_+_30034931
|
0.443
|
NM_021959
|
PPP1R11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11
|
|
chr20_+_56964162
|
0.442
|
NM_001195677
NM_004738
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chr8_+_104310660
|
0.441
|
NM_001164615
|
FZD6
|
frizzled family receptor 6
|
|
chr3_-_194188967
|
0.441
|
NM_024524
|
ATP13A3
|
ATPase type 13A3
|
|
chr16_-_66785511
|
0.439
|
NM_006141
|
DYNC1LI2
|
dynein, cytoplasmic 1, light intermediate chain 2
|
|
chr3_-_170626425
|
0.438
|
NM_020390
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr3_-_52001388
|
0.438
|
NM_020418
NM_001174100
NM_033008
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr11_-_34379526
|
0.437
|
NM_145804
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2
|
|
chrX_-_41782230
|
0.437
|
NM_001126054
NM_001126055
NM_003688
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family)
|
|
chr16_+_23568833
|
0.436
|
NM_019116
|
UBFD1
|
ubiquitin family domain containing 1
|
|
chrY_+_15016018
|
0.433
|
NM_001122665
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr12_+_6833149
|
0.433
|
NM_001164094
NM_016319
NM_001164095
|
COPS7A
|
COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis)
|
|
chrX_-_133119624
|
0.432
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr5_+_140345746
|
0.431
|
NM_018899
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|