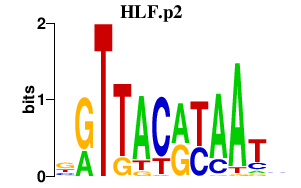
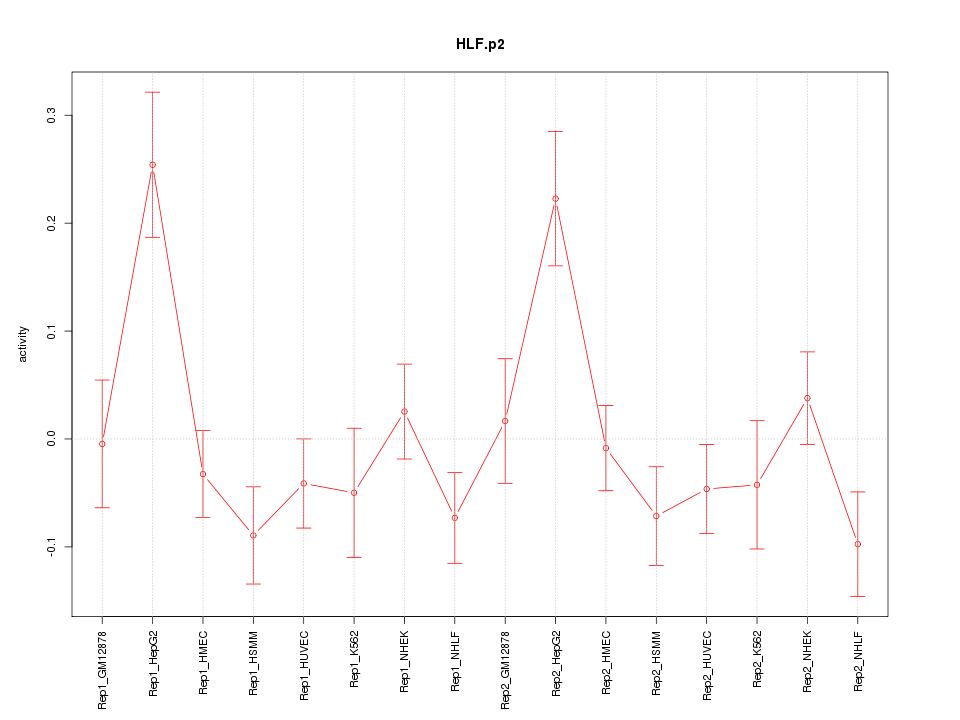
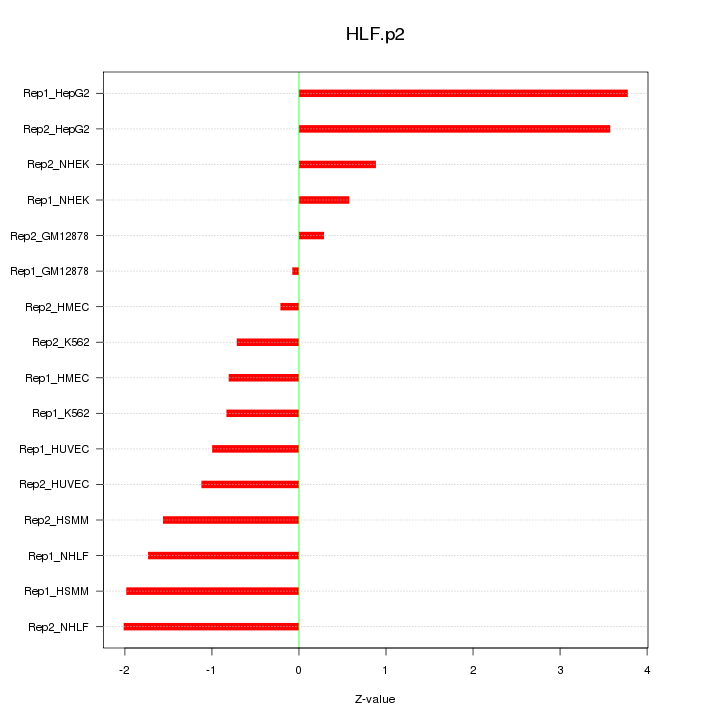
|
chr9_+_117085302
|
7.285
|
NM_000607
|
ORM1
|
orosomucoid 1
|
|
chr9_+_117092068
|
7.198
|
NM_000608
|
ORM2
|
orosomucoid 2
|
|
chr7_+_45933096
|
6.985
|
|
|
|
|
chr10_-_52645430
|
6.879
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr1_-_230849863
|
6.067
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr3_+_133494275
|
5.888
|
|
TF
|
transferrin
|
|
chr16_+_72088507
|
5.413
|
NM_001126102
NM_005143
|
HP
|
haptoglobin
|
|
chr1_-_230849847
|
5.032
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr2_-_88427535
|
5.000
|
NM_001443
|
FABP1
|
fatty acid binding protein 1, liver
|
|
chr16_-_55866972
|
4.821
|
|
CES1
|
carboxylesterase 1
|
|
chr3_+_186330844
|
4.740
|
NM_001622
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr16_-_55867016
|
4.603
|
NM_001025194
NM_001025195
NM_001266
|
CES1
|
carboxylesterase 1
|
|
chr2_+_113885137
|
4.497
|
NM_173842
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr3_-_49726081
|
4.245
|
NM_020998
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like)
|
|
chr2_-_99279935
|
3.883
|
NM_001160154
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr1_+_207262211
|
3.636
|
NM_001017365
NM_001017366
|
C4BPB
|
complement component 4 binding protein, beta
|
|
chr19_+_35773409
|
3.563
|
NM_021175
|
HAMP
|
hepcidin antimicrobial peptide
|
|
chr10_-_95360959
|
3.274
|
NM_006744
|
RBP4
|
retinol binding protein 4, plasma
|
|
chr3_+_186330859
|
3.214
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr13_-_46679143
|
3.160
|
NM_001872
NM_016413
|
CPB2
|
carboxypeptidase B2 (plasma)
|
|
chr4_+_100485239
|
3.111
|
NM_000253
|
MTTP
|
microsomal triglyceride transfer protein
|
|
chr16_+_72097124
|
2.954
|
NM_020995
|
HPR
|
haptoglobin-related protein
|
|
chr4_+_155484131
|
2.748
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr2_+_113885145
|
2.716
|
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr16_+_57406398
|
2.715
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr6_+_131894343
|
2.608
|
NM_000045
NM_001244438
|
ARG1
|
arginase, liver
|
|
chr18_+_55888799
|
2.568
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr17_-_41623246
|
2.457
|
NM_001986
|
ETV4
|
ets variant 4
|
|
chr3_-_120400957
|
2.442
|
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr18_+_55888750
|
2.421
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr6_+_31895201
|
2.368
|
NM_000063
NM_001145903
|
C2
|
complement component 2
|
|
chr10_-_116444374
|
2.357
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr3_+_186330711
|
2.206
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr17_-_34328607
|
2.194
|
|
CCL14-CCL15
|
CCL14-CCL15 readthrough
|
|
chr10_-_116444112
|
2.151
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr3_-_12586962
|
2.085
|
NM_001195279
|
LOC100129480
|
uncharacterized LOC100129480
|
|
chr4_+_74269971
|
2.029
|
NM_000477
|
ALB
|
albumin
|
|
chr9_+_103947316
|
2.012
|
NM_017753
|
LPPR1
|
lipid phosphate phosphatase-related protein type 1
|
|
chr18_+_55816546
|
1.976
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr17_-_34328530
|
1.963
|
|
CCL14-CCL15
CCL15
|
CCL14-CCL15 readthrough
chemokine (C-C motif) ligand 15
|
|
chr17_-_34329083
|
1.959
|
NM_032965
|
CCL15
|
chemokine (C-C motif) ligand 15
|
|
chr17_-_64225496
|
1.951
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr14_+_24563369
|
1.894
|
NM_001018073
NM_004563
|
PCK2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial)
|
|
chr11_-_18258383
|
1.839
|
NM_006512
|
SAA4
|
serum amyloid A4, constitutive
|
|
chr15_-_42840910
|
1.836
|
|
LRRC57
|
leucine rich repeat containing 57
|
|
chr17_-_39092714
|
1.796
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr8_+_40010986
|
1.760
|
NM_020130
|
C8orf4
|
chromosome 8 open reading frame 4
|
|
chr4_+_69962191
|
1.742
|
NM_001074
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7
|
|
chr5_+_150399980
|
1.688
|
NM_002084
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr10_+_114133775
|
1.675
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr6_+_31895401
|
1.650
|
|
C2
|
complement component 2
|
|
chr10_-_82049178
|
1.648
|
NM_000429
|
MAT1A
|
methionine adenosyltransferase I, alpha
|
|
chr1_-_230850335
|
1.593
|
NM_000029
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr20_+_36011936
|
1.559
|
|
SRC
|
v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian)
|
|
chr22_+_24979717
|
1.553
|
NM_013430
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr10_+_124151818
|
1.537
|
NM_021622
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr18_+_55888838
|
1.503
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr8_+_124194751
|
1.440
|
NM_032899
NM_207006
|
FAM83A
|
family with sequence similarity 83, member A
|
|
chr2_-_114514196
|
1.373
|
NM_025181
|
SLC35F5
|
solute carrier family 35, member F5
|
|
chr7_+_129847699
|
1.351
|
NM_145268
|
C7orf45
|
chromosome 7 open reading frame 45
|
|
chr12_-_123201319
|
1.325
|
NM_006018
|
HCAR3
|
hydroxycarboxylic acid receptor 3
|
|
chr4_+_71062241
|
1.292
|
NM_017855
|
ODAM
|
odontogenic, ameloblast asssociated
|
|
chr16_-_4588738
|
1.269
|
NM_001199055
NM_001199056
NM_013399
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr2_+_228678556
|
1.268
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr9_-_94186140
|
1.259
|
NM_005384
|
NFIL3
|
nuclear factor, interleukin 3 regulated
|
|
chr22_+_25003610
|
1.251
|
NM_001032365
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr19_+_45417920
|
1.236
|
NM_001645
|
APOC1
|
apolipoprotein C-I
|
|
chr4_-_110723334
|
1.189
|
NM_000204
|
CFI
|
complement factor I
|
|
chr12_-_8043743
|
1.185
|
|
SLC2A14
|
solute carrier family 2 (facilitated glucose transporter), member 14
|
|
chr1_+_150954498
|
1.182
|
NM_003568
|
ANXA9
|
annexin A9
|
|
chr3_+_142342265
|
1.176
|
NM_002670
|
PLS1
|
plastin 1
|
|
chr6_+_144612872
|
1.161
|
NM_007124
|
UTRN
|
utrophin
|
|
chr19_+_45418063
|
1.154
|
|
APOC1
|
apolipoprotein C-I
|
|
chr4_+_187187342
|
1.147
|
|
F11
|
coagulation factor XI
|
|
chr3_-_170744458
|
1.119
|
|
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2
|
|
chr9_-_130712866
|
1.115
|
NM_203305
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr20_-_3154169
|
1.060
|
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr1_+_229440128
|
1.042
|
NM_006542
|
SPHAR
|
S-phase response (cyclin related)
|
|
chr3_-_99594915
|
1.041
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr9_+_71819926
|
1.036
|
NM_001170415
NM_001170416
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr7_+_94297448
|
1.029
|
|
PEG10
|
paternally expressed 10
|
|
chr18_+_77866914
|
1.022
|
NM_014913
|
ADNP2
|
ADNP homeobox 2
|
|
chr15_+_42840991
|
1.019
|
NM_001130447
NM_018097
|
HAUS2
|
HAUS augmin-like complex, subunit 2
|
|
chr15_-_42840992
|
0.980
|
NM_153260
|
LRRC57
|
leucine rich repeat containing 57
|
|
chr11_-_66675334
|
0.969
|
NM_022172
|
PC
|
pyruvate carboxylase
|
|
chr4_-_23891608
|
0.930
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr3_+_122044010
|
0.928
|
NM_005213
|
CSTA
|
cystatin A (stefin A)
|
|
chr3_+_119501556
|
0.926
|
NM_022002
|
NR1I2
|
nuclear receptor subfamily 1, group I, member 2
|
|
chr19_-_460995
|
0.913
|
NM_012435
|
SHC2
|
SHC (Src homology 2 domain containing) transforming protein 2
|
|
chr14_-_20801427
|
0.882
|
NM_021178
NM_182849
|
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase
|
|
chr19_+_45971249
|
0.881
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr12_-_123187842
|
0.864
|
NM_177551
|
HCAR2
|
hydroxycarboxylic acid receptor 2
|
|
chr16_+_3507956
|
0.843
|
NM_001083600
NM_024845
|
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit
|
|
chr10_+_114133838
|
0.836
|
NM_203379
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr6_-_133035180
|
0.814
|
NM_004666
|
VNN1
|
vanin 1
|
|
chrX_-_131547536
|
0.804
|
NM_001170701
NM_001170702
NM_001170703
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr10_-_101190201
|
0.798
|
NM_002079
|
GOT1
|
glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1)
|
|
chr9_-_99382073
|
0.795
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr8_+_117778741
|
0.794
|
NM_032334
|
UTP23
|
UTP23, small subunit (SSU) processome component, homolog (yeast)
|
|
chr19_+_33864574
|
0.791
|
NM_001806
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma
|
|
chr7_+_20655244
|
0.787
|
NM_001163941
|
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5
|
|
chr3_+_124303505
|
0.783
|
NM_007064
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr11_-_70672644
|
0.774
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr5_+_150400132
|
0.770
|
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr4_-_74088773
|
0.768
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr17_-_56494930
|
0.767
|
NM_017763
|
RNF43
|
ring finger protein 43
|
|
chr3_-_148939784
|
0.757
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr16_+_31711914
|
0.744
|
|
KIAA0664L3
|
KIAA0664-like 3
|
|
chr3_+_169684549
|
0.731
|
NM_003262
|
SEC62
|
SEC62 homolog (S. cerevisiae)
|
|
chr8_-_145115565
|
0.727
|
NM_017570
|
OPLAH
|
5-oxoprolinase (ATP-hydrolysing)
|
|
chr17_-_71308118
|
0.727
|
NM_012121
|
CDC42EP4
|
CDC42 effector protein (Rho GTPase binding) 4
|
|
chr7_+_143880547
|
0.721
|
NM_198495
|
CTAGE4
CTAGE8
|
CTAGE family, member 4
CTAGE family, member 8
|
|
chr18_+_77867145
|
0.714
|
|
ADNP2
|
ADNP homeobox 2
|
|
chr5_-_147211141
|
0.713
|
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1
|
|
chr19_-_2042022
|
0.703
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr11_-_70507911
|
0.683
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr1_+_168148215
|
0.682
|
|
TIPRL
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae)
|
|
chr4_-_110723127
|
0.669
|
|
CFI
|
complement factor I
|
|
chr2_-_73869493
|
0.669
|
NM_003960
|
NAT8
|
N-acetyltransferase 8 (GCN5-related, putative)
|
|
chr17_-_7155258
|
0.657
|
NM_015343
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1
|
|
chr6_-_41702734
|
0.656
|
NM_007162
|
TFEB
|
transcription factor EB
|
|
chr6_-_30043547
|
0.655
|
NM_170769
NM_025236
|
RNF39
|
ring finger protein 39
|
|
chrM_+_1670
|
0.644
|
|
|
|
|
chrX_+_43514157
|
0.643
|
|
MAOA
|
monoamine oxidase A
|
|
chr7_-_143966380
|
0.630
|
NM_198495
|
CTAGE4
CTAGE8
|
CTAGE family, member 4
CTAGE family, member 8
|
|
chr10_+_5238756
|
0.629
|
NM_001818
|
AKR1C4
|
aldo-keto reductase family 1, member C4 (chlordecone reductase; 3-alpha hydroxysteroid dehydrogenase, type I; dihydrodiol dehydrogenase 4)
|
|
chr21_-_43187197
|
0.602
|
NM_020639
|
RIPK4
|
receptor-interacting serine-threonine kinase 4
|
|
chr2_+_201756639
|
0.599
|
NM_001142356
|
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae)
|
|
chr14_+_68086655
|
0.599
|
|
ARG2
|
arginase, type II
|
|
chr6_-_132032156
|
0.599
|
NM_001145659
|
CTAGE9
|
CTAGE family, member 9
|
|
chr5_-_32388595
|
0.598
|
|
ZFR
|
zinc finger RNA binding protein
|
|
chr16_+_31128973
|
0.596
|
NM_032188
NM_182958
|
KAT8
|
K(lysine) acetyltransferase 8
|
|
chr13_-_75901897
|
0.588
|
|
TBC1D4
|
TBC1 domain family, member 4
|
|
chr16_+_31711926
|
0.587
|
|
KIAA0664L3
|
KIAA0664-like 3
|
|
chr21_-_43786602
|
0.587
|
NM_003225
|
TFF1
|
trefoil factor 1
|
|
chr19_-_4540078
|
0.585
|
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1
|
|
chr19_+_13056671
|
0.578
|
|
RAD23A
|
RAD23 homolog A (S. cerevisiae)
|
|
chr21_+_45079375
|
0.575
|
NM_015056
|
RRP1B
|
ribosomal RNA processing 1 homolog B (S. cerevisiae)
|
|
chr12_+_57623355
|
0.573
|
NM_001166356
NM_005412
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial)
|
|
chr1_+_241695433
|
0.573
|
NM_003679
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase)
|
|
chr2_-_239197091
|
0.569
|
NM_022817
|
PER2
|
period homolog 2 (Drosophila)
|
|
chr14_+_103388992
|
0.565
|
NM_030943
|
AMN
|
amnionless homolog (mouse)
|
|
chr17_-_26941178
|
0.552
|
NM_001174103
|
SGK494
|
uncharacterized serine/threonine-protein kinase SgK494
|
|
chr12_-_28124915
|
0.546
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr11_+_43665772
|
0.546
|
|
|
|
|
chr19_-_47617008
|
0.544
|
NM_015168
|
ZC3H4
|
zinc finger CCCH-type containing 4
|
|
chr19_+_13056607
|
0.538
|
NM_005053
|
RAD23A
|
RAD23 homolog A (S. cerevisiae)
|
|
chr3_+_142375738
|
0.537
|
NM_001172312
|
PLS1
|
plastin 1
|
|
chr1_+_212738675
|
0.532
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr17_-_61850953
|
0.525
|
NM_020198
|
CCDC47
|
coiled-coil domain containing 47
|
|
chr11_+_62104773
|
0.523
|
NM_001083926
NM_025080
|
ASRGL1
|
asparaginase like 1
|
|
chr5_+_6746100
|
0.515
|
NM_001171806
|
PAPD7
|
PAP associated domain containing 7
|
|
chr6_+_32939974
|
0.515
|
|
BRD2
|
bromodomain containing 2
|
|
chr5_-_42811959
|
0.515
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr17_+_41322413
|
0.514
|
NM_031862
NM_031858
|
NBR1
|
neighbor of BRCA1 gene 1
|
|
chr16_-_30457197
|
0.511
|
NM_012248
|
SEPHS2
|
selenophosphate synthetase 2
|
|
chr12_+_57623827
|
0.503
|
NM_001166358
NM_001166359
NM_001166357
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial)
|
|
chr17_-_4607414
|
0.497
|
|
PELP1
|
proline, glutamate and leucine rich protein 1
|
|
chr4_-_153273683
|
0.491
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr6_-_2840683
|
0.489
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr3_+_186435097
|
0.484
|
NM_000893
NM_001102416
NM_001166451
|
KNG1
|
kininogen 1
|
|
chr20_-_44600939
|
0.484
|
|
ZNF335
|
zinc finger protein 335
|
|
chr20_+_32150139
|
0.482
|
NM_001039709
NM_005093
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr2_-_68694389
|
0.480
|
NM_001024680
|
FBXO48
|
F-box protein 48
|
|
chr17_-_76836852
|
0.472
|
NM_025090
|
USP36
|
ubiquitin specific peptidase 36
|
|
chr14_+_29241989
|
0.470
|
|
C14orf23
|
chromosome 14 open reading frame 23
|
|
chr7_-_15601574
|
0.468
|
NM_001004320
|
AGMO
|
alkylglycerol monooxygenase
|
|
chr7_+_143268872
|
0.462
|
NM_001008747
|
CTAGE6P
CTAGE15P
|
CTAGE family, member 6, pseudogene
CTAGE family, member 15, pseudogene
|
|
chr3_+_42132707
|
0.458
|
NM_001042646
|
TRAK1
|
trafficking protein, kinesin binding 1
|
|
chr1_-_154600383
|
0.457
|
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr1_+_179965756
|
0.455
|
|
CEP350
|
centrosomal protein 350kDa
|
|
chr5_+_156696351
|
0.454
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr22_+_35462125
|
0.450
|
NM_001008494
|
ISX
|
intestine-specific homeobox
|
|
chr17_-_4607198
|
0.445
|
|
PELP1
|
proline, glutamate and leucine rich protein 1
|
|
chr20_+_62372436
|
0.445
|
|
SLC2A4RG
|
SLC2A4 regulator
|
|
chrX_+_9431314
|
0.445
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr1_-_247054277
|
0.439
|
|
AHCTF1
|
AT hook containing transcription factor 1
|
|
chr1_-_247094659
|
0.436
|
NM_015446
|
AHCTF1
AHCTF1P1
|
AT hook containing transcription factor 1
AT hook containing transcription factor 1 pseudogene 1
|
|
chr6_-_47254143
|
0.433
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr17_-_61850890
|
0.428
|
|
CCDC47
|
coiled-coil domain containing 47
|
|
chr1_-_154600446
|
0.427
|
NM_001025107
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr2_-_214015053
|
0.425
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr10_+_71812536
|
0.421
|
|
H2AFY2
|
H2A histone family, member Y2
|
|
chr8_+_21916685
|
0.414
|
NM_001114138
NM_001978
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr2_-_216300770
|
0.412
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr3_+_9975513
|
0.412
|
NM_015513
NM_001031717
NM_001077415
|
CRELD1
|
cysteine-rich with EGF-like domains 1
|
|
chr19_-_4540035
|
0.412
|
NM_052972
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1
|
|
chr2_-_31637598
|
0.410
|
NM_000379
|
XDH
|
xanthine dehydrogenase
|
|
chr1_-_200638996
|
0.407
|
|
DDX59
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 59
|
|
chr8_-_101315457
|
0.405
|
NM_015435
|
RNF19A
|
ring finger protein 19A
|
|
chr2_-_65593866
|
0.403
|
NM_001128210
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr12_-_7261788
|
0.403
|
NM_016546
|
C1RL
|
complement component 1, r subcomponent-like
|
|
chr10_+_103911932
|
0.399
|
NM_004741
|
NOLC1
|
nucleolar and coiled-body phosphoprotein 1
|
|
chr19_+_13056656
|
0.399
|
|
RAD23A
|
RAD23 homolog A (S. cerevisiae)
|
|
chr19_-_6459735
|
0.399
|
NM_024103
|
SLC25A23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23
|
|
chr4_+_1894508
|
0.398
|
NM_007331
NM_133330
NM_133331
NM_133335
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr5_+_126112314
|
0.392
|
NM_005573
|
LMNB1
|
lamin B1
|
|
chr9_-_112191105
|
0.385
|
NM_001145371
NM_001145372
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr10_-_45474210
|
0.383
|
NM_007021
|
C10orf10
|
chromosome 10 open reading frame 10
|
|
chr16_-_70323343
|
0.378
|
|
AARS
|
alanyl-tRNA synthetase
|