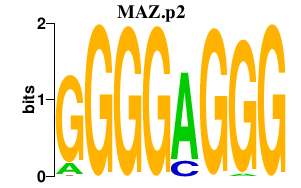
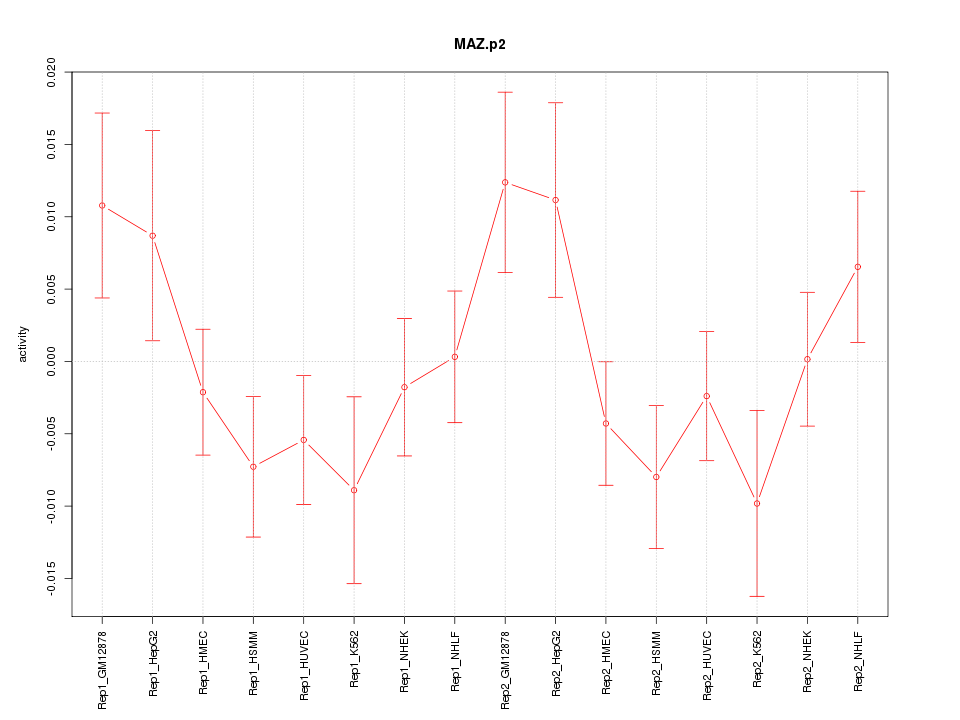
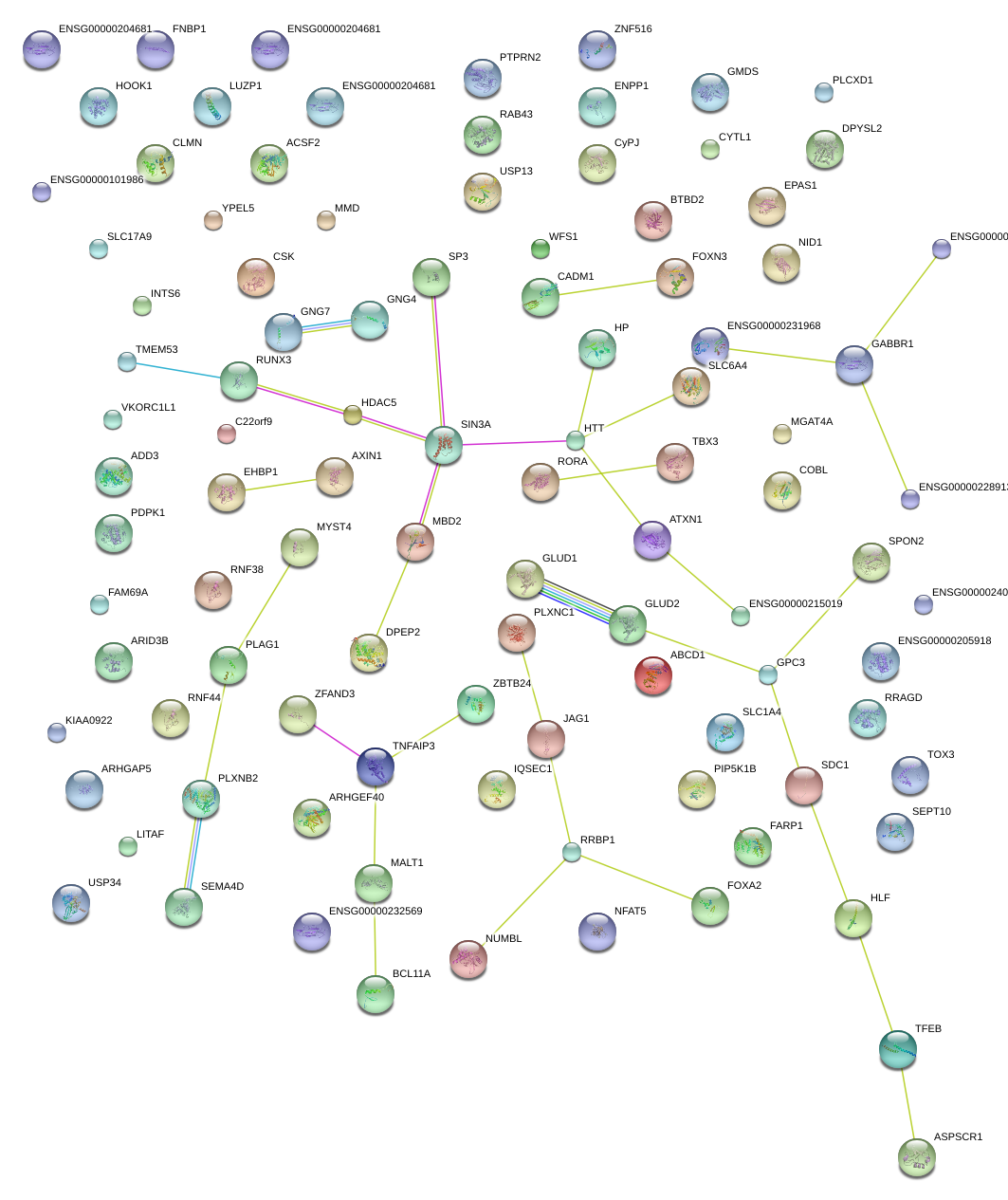
|
chr9_-_132805456
|
2.211
|
NM_015033
|
FNBP1
|
formin binding protein 1
|
|
chrX_+_198060
|
2.169
|
NM_018390
|
PLCXD1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1
|
|
chr19_-_41196419
|
2.061
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|
|
chr6_-_29595746
|
1.854
|
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr9_-_132805256
|
1.788
|
|
FNBP1
|
formin binding protein 1
|
|
chr16_-_11680756
|
1.736
|
NM_001136472
NM_001136473
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr8_+_26371461
|
1.696
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chrY_+_148060
|
1.664
|
NM_018390
|
PLCXD1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1
|
|
chr14_-_89883306
|
1.645
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr6_-_90121656
|
1.488
|
|
RRAGD
|
Ras-related GTP binding D
|
|
chr10_+_76586299
|
1.410
|
NM_012330
|
KAT6B
|
K(lysine) acetyltransferase 6B
|
|
chr18_+_56338783
|
1.335
|
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1
|
|
chr16_-_52580805
|
1.271
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr7_-_158380360
|
1.229
|
NM_002847
NM_130842
NM_130843
|
PTPRN2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2
|
|
chr18_+_56338617
|
1.192
|
NM_006785
NM_173844
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1
|
|
chr4_-_1166458
|
1.173
|
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr14_-_95786095
|
1.169
|
NM_024734
|
CLMN
|
calmin (calponin-like, transmembrane)
|
|
chrX_-_133119624
|
1.150
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr4_-_1166364
|
1.149
|
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr6_+_37787592
|
1.140
|
|
ZFAND3
|
zinc finger, AN1-type domain 3
|
|
chr3_-_13008959
|
1.135
|
NM_014869
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr18_+_56338678
|
1.111
|
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1
|
|
chr2_-_99347430
|
1.080
|
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr19_-_2702662
|
1.071
|
|
GNG7
|
guanine nucleotide binding protein (G protein), gamma 7
|
|
chr16_+_69600110
|
1.058
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr18_+_56338818
|
1.050
|
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1
|
|
chr2_-_110371669
|
1.049
|
NM_144710
NM_178584
|
SEPT10
|
septin 10
|
|
chr17_-_42201009
|
1.045
|
NM_001015053
NM_005474
|
HDAC5
|
histone deacetylase 5
|
|
chr4_-_1166547
|
1.013
|
NM_012445
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr2_-_60780767
|
1.011
|
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr6_-_41702734
|
0.992
|
NM_007162
|
TFEB
|
transcription factor EB
|
|
chr2_-_60780404
|
0.986
|
NM_018014
NM_022893
NM_138559
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr16_+_69599869
|
0.978
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr4_-_1166378
|
0.967
|
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr2_-_61697899
|
0.963
|
|
USP34
|
ubiquitin specific peptidase 34
|
|
chr17_+_53342356
|
0.929
|
|
HLF
|
hepatic leukemia factor
|
|
chr3_+_179370779
|
0.904
|
NM_003940
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3)
|
|
chr9_-_36401149
|
0.901
|
NM_194330
|
RNF38
|
ring finger protein 38
|
|
chr17_+_48503518
|
0.888
|
NM_025149
|
ACSF2
|
acyl-CoA synthetase family member 2
|
|
chr1_+_60280462
|
0.883
|
NM_015888
|
HOOK1
|
hook homolog 1 (Drosophila)
|
|
chr1_-_93426998
|
0.876
|
NM_001006605
NM_001252269
NM_001252270
NM_001252273
|
FAM69A
|
family with sequence similarity 69, member A
|
|
chr6_-_90121911
|
0.869
|
|
RRAGD
|
Ras-related GTP binding D
|
|
chr1_-_25256364
|
0.867
|
|
RUNX3
|
runt-related transcription factor 3
|
|
chr17_-_53499055
|
0.862
|
NM_012329
|
MMD
|
monocyte to macrophage differentiation-associated
|
|
chr9_-_92112887
|
0.861
|
|
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D
|
|
chr17_-_42200957
|
0.855
|
|
HDAC5
|
histone deacetylase 5
|
|
chr20_-_22564893
|
0.855
|
|
FOXA2
|
forkhead box A2
|
|
chrX_-_133119475
|
0.849
|
|
GPC3
|
glypican 3
|
|
chr10_-_88854686
|
0.844
|
NM_005271
|
GLUD1
|
glutamate dehydrogenase 1
|
|
chr4_-_1166959
|
0.843
|
NM_001128325
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr6_+_132129195
|
0.842
|
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr6_+_138188352
|
0.842
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr17_+_48503607
|
0.841
|
|
ACSF2
|
acyl-CoA synthetase family member 2
|
|
chr2_+_62932786
|
0.840
|
NM_001142616
NM_015252
|
EHBP1
|
EH domain binding protein 1
|
|
chr14_+_32546445
|
0.835
|
NM_001030055
NM_001173
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr1_-_23495284
|
0.823
|
|
LUZP1
|
leucine zipper protein 1
|
|
chr15_+_75074714
|
0.820
|
|
CSK
|
c-src tyrosine kinase
|
|
chr15_-_60884615
|
0.819
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr10_-_88854490
|
0.819
|
|
GLUD1
|
glutamate dehydrogenase 1
|
|
chr6_-_16761600
|
0.808
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr14_+_21538339
|
0.807
|
NM_018071
|
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40
|
|
chr19_-_2015628
|
0.807
|
NM_017797
|
BTBD2
|
BTB (POZ) domain containing 2
|
|
chr17_+_79935410
|
0.799
|
NM_001251888
NM_024083
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|
|
chr20_-_10654429
|
0.794
|
NM_000214
|
JAG1
|
jagged 1
|
|
chr18_-_74207145
|
0.787
|
NM_014643
|
ZNF516
|
zinc finger protein 516
|
|
chr3_-_128840343
|
0.778
|
|
RAB43
|
RAB43, member RAS oncogene family
|
|
chr17_-_79008372
|
0.775
|
|
FLJ90757
|
uncharacterized LOC440465
|
|
chr7_-_51384458
|
0.772
|
NM_015198
|
COBL
|
cordon-bleu homolog (mouse)
|
|
chr1_-_25256767
|
0.769
|
NM_004350
|
RUNX3
|
runt-related transcription factor 3
|
|
chr2_-_110371404
|
0.765
|
|
SEPT10
|
septin 10
|
|
chr2_+_65215577
|
0.763
|
NM_001193493
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4
|
|
chr15_+_74833517
|
0.759
|
NM_006465
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like)
|
|
chr15_-_75743776
|
0.747
|
NM_015477
|
SIN3A
|
SIN3 transcription regulator homolog A (yeast)
|
|
chr7_+_65338161
|
0.746
|
NM_173517
|
VKORC1L1
|
vitamin K epoxide reductase complex, subunit 1-like 1
|
|
chr20_-_17662828
|
0.744
|
NM_001042576
NM_004587
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr10_-_88854611
|
0.743
|
|
GLUD1
|
glutamate dehydrogenase 1
|
|
chr16_-_11680146
|
0.740
|
NM_004862
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr2_-_174828775
|
0.734
|
NM_001017371
|
SP3
|
Sp3 transcription factor
|
|
chr4_+_3076236
|
0.730
|
NM_002111
|
HTT
|
huntingtin
|
|
chr4_+_6271588
|
0.727
|
|
WFS1
|
Wolfram syndrome 1 (wolframin)
|
|
chr12_+_94542239
|
0.721
|
NM_005761
|
PLXNC1
|
plexin C1
|
|
chr16_+_69599868
|
0.717
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
NM_173215
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr22_-_50746000
|
0.712
|
NM_012401
|
PLXNB2
|
plexin B2
|
|
chr18_-_51750460
|
0.704
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr6_-_109804432
|
0.689
|
NM_001164313
NM_014797
|
ZBTB24
|
zinc finger and BTB domain containing 24
|
|
chr11_-_115375065
|
0.688
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr2_-_20424627
|
0.687
|
NM_002997
|
SDC1
|
syndecan 1
|
|
chr6_-_2245655
|
0.682
|
NM_001500
|
GMDS
|
GDP-mannose 4,6-dehydratase
|
|
chr12_-_115121939
|
0.678
|
NM_005996
NM_016569
|
TBX3
|
T-box 3
|
|
chrX_+_152990322
|
0.676
|
NM_000033
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1
|
|
chr16_-_402617
|
0.674
|
NM_003502
NM_181050
|
AXIN1
|
axin 1
|
|
chr13_-_52027133
|
0.669
|
|
INTS6
|
integrator complex subunit 6
|
|
chr16_+_2588000
|
0.669
|
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr4_+_154387450
|
0.666
|
NM_001131007
NM_015196
|
KIAA0922
|
KIAA0922
|
|
chr8_-_57123827
|
0.665
|
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr5_-_175964223
|
0.663
|
NM_014901
|
RNF44
|
ring finger protein 44
|
|
chr22_-_45636584
|
0.654
|
NM_001009880
|
KIAA0930
|
KIAA0930
|
|
chr20_-_17662704
|
0.651
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr13_+_98795710
|
0.650
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr22_-_45636760
|
0.648
|
|
KIAA0930
|
KIAA0930
|
|
chr2_+_30369868
|
0.643
|
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr16_+_2587989
|
0.638
|
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr1_-_236228476
|
0.636
|
NM_002508
|
NID1
|
nidogen 1
|
|
chr9_+_71320105
|
0.633
|
NM_003558
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta
|
|
chr1_-_45140117
|
0.632
|
|
TMEM53
|
transmembrane protein 53
|
|
chr10_+_111767877
|
0.630
|
|
ADD3
|
adducin 3 (gamma)
|
|
chr10_-_88854595
|
0.623
|
|
GLUD1
|
glutamate dehydrogenase 1
|
|
chr1_-_235812994
|
0.620
|
NM_001098722
|
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4
|
|
chr20_+_61584094
|
0.620
|
|
SLC17A9
|
solute carrier family 17, member 9
|
|
chr17_+_47653474
|
0.613
|
|
NXPH3
|
neurexophilin 3
|
|
chr20_+_35201948
|
0.613
|
NM_021809
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr20_+_57466721
|
0.612
|
|
GNAS
|
GNAS complex locus
|
|
chr1_+_1981902
|
0.611
|
NM_002744
|
PRKCZ
|
protein kinase C, zeta
|
|
chr12_+_53846086
|
0.607
|
|
PCBP2
|
poly(rC) binding protein 2
|
|
chr11_-_46142955
|
0.605
|
NM_001101802
NM_016621
|
PHF21A
|
PHD finger protein 21A
|
|
chr1_-_151431892
|
0.597
|
NM_001194937
NM_001194938
NM_015100
NM_145796
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr10_-_88854568
|
0.597
|
|
GLUD1
|
glutamate dehydrogenase 1
|
|
chr1_-_11714738
|
0.595
|
NM_012168
|
FBXO2
|
F-box protein 2
|
|
chr10_-_88854645
|
0.595
|
|
GLUD1
|
glutamate dehydrogenase 1
|
|
chr6_+_107811272
|
0.593
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr16_+_8768403
|
0.593
|
NM_020686
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr4_+_6271605
|
0.589
|
|
WFS1
|
Wolfram syndrome 1 (wolframin)
|
|
chr2_+_54683276
|
0.588
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr9_-_36400817
|
0.587
|
NM_194328
NM_194332
|
RNF38
|
ring finger protein 38
|
|
chr5_-_111093251
|
0.585
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr10_+_45869667
|
0.584
|
|
ALOX5
|
arachidonate 5-lipoxygenase
|
|
chr6_-_31239489
|
0.584
|
|
HLA-B
HLA-C
|
major histocompatibility complex, class I, B
major histocompatibility complex, class I, C
|
|
chr14_-_38064176
|
0.581
|
|
FOXA1
|
forkhead box A1
|
|
chrX_-_19002444
|
0.581
|
NM_000292
|
PHKA2
|
phosphorylase kinase, alpha 2 (liver)
|
|
chr9_+_2622099
|
0.581
|
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr18_+_11981421
|
0.578
|
NM_014214
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2
|
|
chr12_+_112204690
|
0.578
|
NM_000690
NM_001204889
|
ALDH2
|
aldehyde dehydrogenase 2 family (mitochondrial)
|
|
chr2_+_79740059
|
0.576
|
NM_001164883
NM_004389
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr8_+_144635556
|
0.574
|
NM_001166237
|
GSDMD
|
gasdermin D
|
|
chr2_-_96811138
|
0.571
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr20_+_61448391
|
0.570
|
NM_001853
|
COL9A3
|
collagen, type IX, alpha 3
|
|
chr19_+_8274179
|
0.568
|
NM_024552
|
CERS4
|
ceramide synthase 4
|
|
chr8_-_103666018
|
0.567
|
NM_001032282
|
KLF10
|
Kruppel-like factor 10
|
|
chr14_-_96180402
|
0.566
|
NM_001098725
NM_021966
|
TCL1A
|
T-cell leukemia/lymphoma 1A
|
|
chr2_+_64681270
|
0.566
|
NM_014181
|
LGALSL
|
lectin, galactoside-binding-like
|
|
chr20_+_35974458
|
0.564
|
NM_198291
|
SRC
|
v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian)
|
|
chr3_+_5021096
|
0.564
|
NM_003670
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr7_-_24797031
|
0.563
|
|
DFNA5
|
deafness, autosomal dominant 5
|
|
chr20_+_388941
|
0.562
|
|
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1
|
|
chr17_+_79935493
|
0.557
|
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|
|
chr9_+_2622052
|
0.557
|
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr19_-_7293876
|
0.557
|
NM_000208
NM_001079817
|
INSR
|
insulin receptor
|
|
chr14_+_60715986
|
0.557
|
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A
|
|
chr1_+_211432855
|
0.553
|
|
RCOR3
|
REST corepressor 3
|
|
chr9_+_2622134
|
0.553
|
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr20_+_388717
|
0.549
|
|
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1
|
|
chr7_-_24797052
|
0.548
|
NM_001127453
|
DFNA5
|
deafness, autosomal dominant 5
|
|
chr6_+_37137882
|
0.546
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr20_-_10654139
|
0.545
|
|
JAG1
|
jagged 1
|
|
chr2_-_174829108
|
0.544
|
|
SP3
|
Sp3 transcription factor
|
|
chr6_+_1390068
|
0.544
|
NM_001452
|
FOXF2
|
forkhead box F2
|
|
chr9_+_112810877
|
0.544
|
NM_001004065
NM_001198656
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr10_+_45869611
|
0.542
|
NM_000698
|
ALOX5
|
arachidonate 5-lipoxygenase
|
|
chr2_+_42396392
|
0.539
|
NM_001145076
NM_019063
|
EML4
|
echinoderm microtubule associated protein like 4
|
|
chr10_+_89623091
|
0.538
|
NM_000314
|
PTEN
|
phosphatase and tensin homolog
|
|
chr20_+_35201854
|
0.532
|
NM_001199513
NM_001199514
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr14_-_38064440
|
0.531
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr9_-_88356800
|
0.526
|
NM_015239
|
AGTPBP1
|
ATP/GTP binding protein 1
|
|
chr6_-_111804106
|
0.524
|
|
REV3L
|
REV3-like, catalytic subunit of DNA polymerase zeta (yeast)
|
|
chr1_+_151584665
|
0.524
|
|
SNX27
|
sorting nexin family member 27
|
|
chr1_+_151584514
|
0.524
|
NM_030918
|
SNX27
|
sorting nexin family member 27
|
|
chr2_+_110371887
|
0.521
|
NM_023016
|
ANKRD57
|
ankyrin repeat domain 57
|
|
chr18_+_60190657
|
0.519
|
NM_017742
|
ZCCHC2
|
zinc finger, CCHC domain containing 2
|
|
chr2_+_64681610
|
0.515
|
|
LGALSL
|
lectin, galactoside-binding-like
|
|
chr7_-_102257199
|
0.515
|
NM_001079877
NM_006989
|
RASA4
|
RAS p21 protein activator 4
|
|
chr2_-_61697846
|
0.512
|
NM_014709
|
USP34
|
ubiquitin specific peptidase 34
|
|
chr16_+_19179534
|
0.512
|
NM_016524
|
SYT17
|
synaptotagmin XVII
|
|
chr12_+_11802642
|
0.511
|
|
ETV6
|
ets variant 6
|
|
chr4_+_6271558
|
0.511
|
NM_001145853
NM_006005
|
WFS1
|
Wolfram syndrome 1 (wolframin)
|
|
chr17_-_47439326
|
0.511
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr19_-_2702738
|
0.510
|
NM_052847
|
GNG7
|
guanine nucleotide binding protein (G protein), gamma 7
|
|
chr14_-_21566497
|
0.510
|
|
ZNF219
|
zinc finger protein 219
|
|
chr1_+_200993041
|
0.510
|
|
|
|
|
chr9_+_137218324
|
0.510
|
|
RXRA
|
retinoid X receptor, alpha
|
|
chr3_-_149688638
|
0.509
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr1_+_11751778
|
0.509
|
NM_198545
|
C1orf187
|
chromosome 1 open reading frame 187
|
|
chr5_+_130599770
|
0.508
|
|
CDC42SE2
|
CDC42 small effector 2
|
|
chr2_+_238600878
|
0.507
|
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr7_+_69064316
|
0.507
|
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr8_-_57123858
|
0.503
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr10_-_97050671
|
0.501
|
NM_020992
|
PDLIM1
|
PDZ and LIM domain 1
|
|
chr1_+_27022521
|
0.498
|
NM_006015
NM_139135
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr2_+_30369749
|
0.497
|
NM_001127399
NM_001127400
NM_001127401
NM_016061
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr20_-_33413368
|
0.497
|
NM_001242539
NM_014071
|
NCOA6
|
nuclear receptor coactivator 6
|
|
chr19_+_18208596
|
0.492
|
NM_015016
|
MAST3
|
microtubule associated serine/threonine kinase 3
|
|
chr4_-_105416050
|
0.492
|
|
CXXC4
|
CXXC finger protein 4
|
|
chr16_-_29910340
|
0.491
|
NM_001114099
NM_001114100
NM_001243332
NM_001243333
NM_012410
NM_201575
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2
|
|
chr17_-_56595192
|
0.489
|
NM_004687
|
MTMR4
|
myotubularin related protein 4
|
|
chr12_-_56652118
|
0.487
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr2_+_46769892
|
0.487
|
|
RHOQ
|
ras homolog gene family, member Q
|
|
chr8_-_99837768
|
0.486
|
NM_006281
|
STK3
|
serine/threonine kinase 3
|
|
chr9_-_6645564
|
0.485
|
NM_000170
|
GLDC
|
glycine dehydrogenase (decarboxylating)
|
|
chr1_-_151431648
|
0.485
|
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr2_-_174828515
|
0.484
|
|
SP3
|
Sp3 transcription factor
|
|
chr5_-_142783059
|
0.483
|
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|