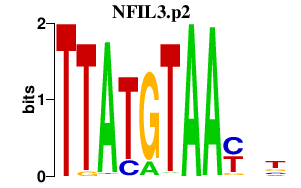
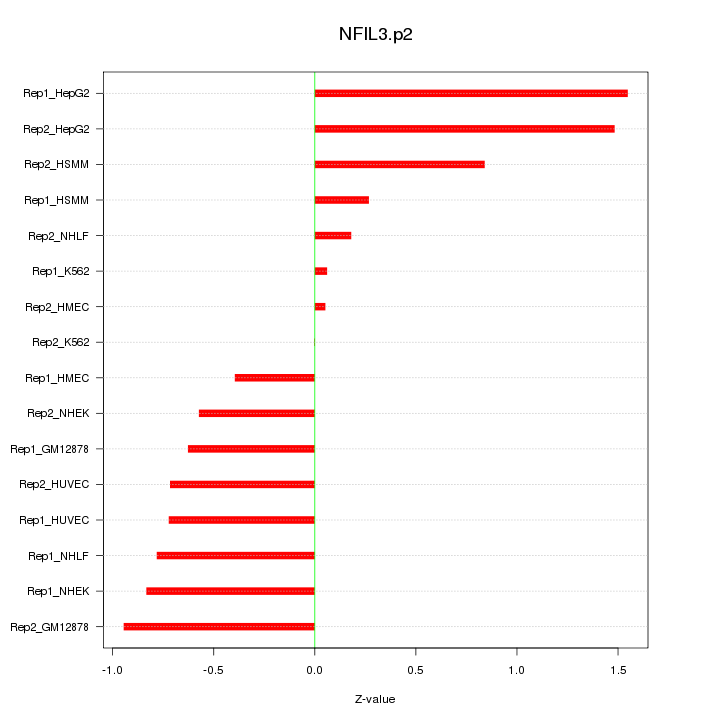
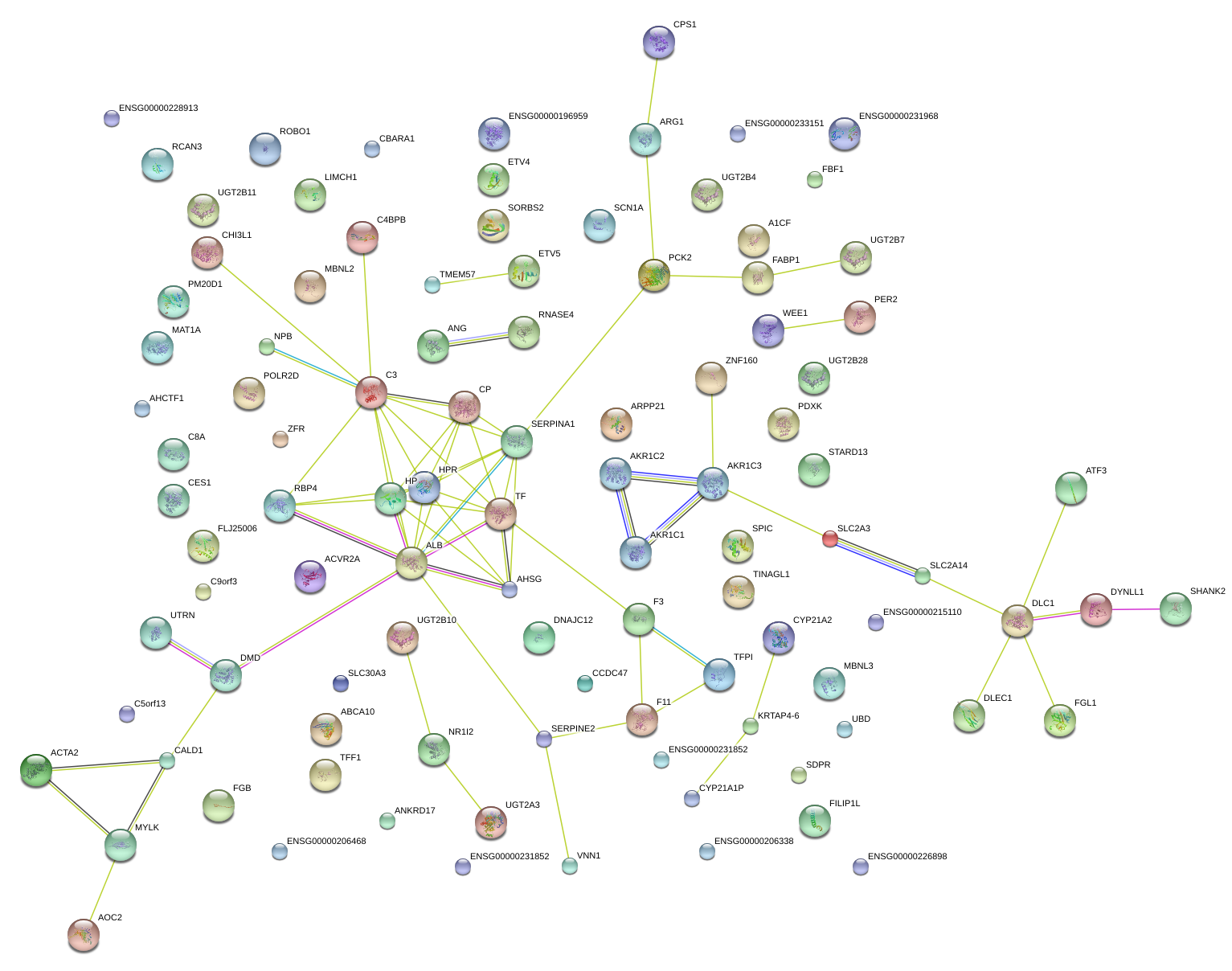
|
chr10_-_95360959
|
6.465
|
NM_006744
|
RBP4
|
retinol binding protein 4, plasma
|
|
chr4_+_74269971
|
6.114
|
NM_000477
|
ALB
|
albumin
|
|
chr3_+_133494275
|
4.363
|
|
TF
|
transferrin
|
|
chr10_-_52645430
|
2.037
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr10_-_82049178
|
1.633
|
NM_000429
|
MAT1A
|
methionine adenosyltransferase I, alpha
|
|
chr14_+_24563369
|
1.590
|
NM_001018073
NM_004563
|
PCK2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial)
|
|
chr8_-_17752847
|
1.541
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr3_+_186330844
|
1.508
|
NM_001622
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr17_-_41623246
|
1.447
|
NM_001986
|
ETV4
|
ets variant 4
|
|
chr1_+_207262211
|
1.414
|
NM_001017365
NM_001017366
|
C4BPB
|
complement component 4 binding protein, beta
|
|
chr17_-_41623652
|
1.381
|
NM_001079675
|
ETV4
|
ets variant 4
|
|
chrX_-_131547536
|
1.334
|
NM_001170701
NM_001170702
NM_001170703
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr6_-_133035180
|
1.254
|
NM_004666
|
VNN1
|
vanin 1
|
|
chr10_-_69597768
|
1.218
|
NM_021800
NM_201262
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr10_+_5005601
|
1.143
|
|
AKR1C1
AKR1C3
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr4_+_155484131
|
1.098
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr10_+_5005453
|
1.006
|
NM_001353
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr2_-_167005641
|
0.983
|
NM_001202435
|
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit
|
|
chr16_-_55866972
|
0.943
|
|
CES1
|
carboxylesterase 1
|
|
chr3_+_186330859
|
0.920
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr3_-_99833348
|
0.875
|
NM_001042459
NM_182909
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr14_-_94849552
|
0.869
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr10_-_5045967
|
0.846
|
NM_001135241
|
AKR1C2
|
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chr10_+_5005685
|
0.786
|
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr1_+_57320442
|
0.748
|
NM_000562
|
C8A
|
complement component 8, alpha polypeptide
|
|
chr4_-_186578122
|
0.743
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_+_131894343
|
0.737
|
NM_000045
NM_001244438
|
ARG1
|
arginase, liver
|
|
chr17_-_67240955
|
0.730
|
NM_080282
|
ABCA10
|
ATP-binding cassette, sub-family A (ABC1), member 10
|
|
chr19_-_6720585
|
0.719
|
NM_000064
|
C3
|
complement component 3
|
|
chr2_-_88427535
|
0.715
|
NM_001443
|
FABP1
|
fatty acid binding protein 1, liver
|
|
chr1_+_212738675
|
0.703
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr9_+_97521930
|
0.673
|
NM_001193329
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr16_-_55867016
|
0.655
|
NM_001025194
NM_001025195
NM_001266
|
CES1
|
carboxylesterase 1
|
|
chr5_-_32388595
|
0.643
|
|
ZFR
|
zinc finger RNA binding protein
|
|
chr3_+_186330711
|
0.633
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr1_+_24840800
|
0.613
|
NM_001251980
NM_001251982
NM_001251985
|
RCAN3
|
RCAN family member 3
|
|
chr12_-_8043743
|
0.586
|
|
SLC2A14
|
solute carrier family 2 (facilitated glucose transporter), member 14
|
|
chr17_-_26941178
|
0.565
|
NM_001174103
|
SGK494
|
uncharacterized serine/threonine-protein kinase SgK494
|
|
chr3_-_78666943
|
0.562
|
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr14_+_21156922
|
0.538
|
NM_001097577
NM_194431
|
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5
ribonuclease, RNase A family, 4
|
|
chr17_-_61850953
|
0.525
|
NM_020198
|
CCDC47
|
coiled-coil domain containing 47
|
|
chr3_-_99594915
|
0.516
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr2_-_192711923
|
0.494
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr7_+_134464161
|
0.491
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr7_+_45933096
|
0.483
|
|
|
|
|
chr10_-_74283693
|
0.479
|
NM_001195519
|
MICU1
|
mitochondrial calcium uptake 1
|
|
chr2_-_188419049
|
0.474
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr16_+_31711914
|
0.461
|
|
KIAA0664L3
|
KIAA0664-like 3
|
|
chr17_-_61850890
|
0.448
|
|
CCDC47
|
coiled-coil domain containing 47
|
|
chr4_+_187187342
|
0.448
|
|
F11
|
coagulation factor XI
|
|
chr2_-_239197091
|
0.445
|
NM_022817
|
PER2
|
period homolog 2 (Drosophila)
|
|
chr1_-_203155910
|
0.413
|
NM_001276
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr3_+_119501556
|
0.408
|
NM_022002
|
NR1I2
|
nuclear receptor subfamily 1, group I, member 2
|
|
chr16_+_72088507
|
0.404
|
NM_001126102
NM_005143
|
HP
|
haptoglobin
|
|
chr2_-_224903324
|
0.396
|
NM_001136530
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr4_-_70080448
|
0.387
|
NM_001073
|
UGT2B11
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B11
UDP glucuronosyltransferase 2 family, polypeptide B10
|
|
chr1_-_203155813
|
0.383
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chrX_-_33229428
|
0.383
|
NM_004006
|
DMD
|
dystrophin
|
|
chr1_-_203155867
|
0.375
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr2_-_188419185
|
0.373
|
NM_001032281
NM_006287
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr13_+_97874540
|
0.370
|
NM_144778
NM_207304
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr3_-_148939784
|
0.369
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr4_+_41700736
|
0.365
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr16_+_3507956
|
0.364
|
NM_001083600
NM_024845
|
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit
|
|
chr4_-_69817478
|
0.360
|
NM_024743
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3
|
|
chr1_-_203155827
|
0.359
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr16_+_31711926
|
0.356
|
|
KIAA0664L3
|
KIAA0664-like 3
|
|
chr17_+_40996596
|
0.351
|
NM_001158
NM_009590
|
AOC2
|
amine oxidase, copper containing 2 (retina-specific)
|
|
chr21_-_43786602
|
0.345
|
NM_003225
|
TFF1
|
trefoil factor 1
|
|
chr1_-_203155745
|
0.343
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr21_+_45138973
|
0.327
|
NM_003681
|
PDXK
|
pyridoxal (pyridoxine, vitamin B6) kinase
|
|
chr4_-_74088773
|
0.320
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr2_+_148602569
|
0.319
|
NM_001616
|
ACVR2A
|
activin A receptor, type IIA
|
|
chr1_-_247094659
|
0.303
|
NM_015446
|
AHCTF1
AHCTF1P1
|
AT hook containing transcription factor 1
AT hook containing transcription factor 1 pseudogene 1
|
|
chr1_+_25757381
|
0.302
|
NM_018202
|
TMEM57
|
transmembrane protein 57
|
|
chr3_-_123339178
|
0.294
|
|
MYLK
|
myosin light chain kinase
|
|
chr6_-_29527477
|
0.292
|
|
UBD
|
ubiquitin D
|
|
chr11_+_9596233
|
0.288
|
NM_001143976
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr13_-_33780142
|
0.283
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr12_-_123201319
|
0.279
|
NM_006018
|
HCAR3
|
hydroxycarboxylic acid receptor 3
|
|
chr5_-_111091947
|
0.278
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr11_-_70672644
|
0.278
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr3_-_185826748
|
0.277
|
NM_004454
|
ETV5
|
ets variant 5
|
|
chr2_+_211421322
|
0.272
|
NM_001875
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr16_+_72097124
|
0.270
|
NM_020995
|
HPR
|
haptoglobin-related protein
|
|
chr22_-_50221195
|
0.268
|
|
BRD1
|
bromodomain containing 1
|
|
chr9_+_139871954
|
0.264
|
NM_000954
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chr12_-_123187842
|
0.256
|
NM_177551
|
HCAR2
|
hydroxycarboxylic acid receptor 2
|
|
chr7_+_20686965
|
0.252
|
NM_001163942
NM_001163993
NM_178559
|
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5
|
|
chr18_+_32621299
|
0.244
|
NM_014268
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr19_-_46389307
|
0.242
|
NM_015649
|
IRF2BP1
|
interferon regulatory factor 2 binding protein 1
|
|
chr3_+_23987514
|
0.239
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr15_-_56209305
|
0.239
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr9_+_139872014
|
0.239
|
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chr1_-_182354649
|
0.236
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr5_+_172483285
|
0.234
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr19_-_17414083
|
0.225
|
NM_024527
|
ABHD8
|
abhydrolase domain containing 8
|
|
chr4_+_70146216
|
0.222
|
NM_001207004
NM_053039
|
UGT2B28
|
UDP glucuronosyltransferase 2 family, polypeptide B28
|
|
chr16_+_7382750
|
0.219
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr17_+_61851557
|
0.211
|
NM_007372
NM_203499
|
DDX42
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 42
|
|
chr12_-_125002828
|
0.210
|
|
|
|
|
chr21_-_27423423
|
0.209
|
|
|
|
|
chr9_-_95298251
|
0.207
|
NM_001197295
NM_001197296
NM_001393
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chr5_-_140013285
|
0.201
|
NM_001040021
NM_001174104
NM_001174105
|
CD14
|
CD14 molecule
|
|
chr3_+_73110809
|
0.201
|
NM_018029
|
EBLN2
|
endogenous Bornavirus-like nucleoprotein 2
|
|
chr7_+_20655244
|
0.200
|
NM_001163941
|
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5
|
|
chr2_+_201473942
|
0.199
|
|
AOX1
|
aldehyde oxidase 1
|
|
chr10_+_124151818
|
0.198
|
NM_021622
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr19_-_3985343
|
0.197
|
|
EEF2
|
eukaryotic translation elongation factor 2
|
|
chr6_-_2840683
|
0.194
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr17_-_39092714
|
0.193
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr6_+_167525294
|
0.193
|
NM_004367
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr14_-_20797532
|
0.189
|
NM_182852
NM_182851
|
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase
|
|
chr9_-_73029539
|
0.186
|
NM_001206
|
KLF9
|
Kruppel-like factor 9
|
|
chr1_-_242162364
|
0.184
|
NM_001004343
|
MAP1LC3C
|
microtubule-associated protein 1 light chain 3 gamma
|
|
chr11_-_128894008
|
0.184
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr2_-_99279935
|
0.181
|
NM_001160154
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr1_+_150898814
|
0.180
|
NM_001145415
NM_001243491
NM_012432
|
SETDB1
|
SET domain, bifurcated 1
|
|
chr11_-_62607043
|
0.179
|
|
WDR74
|
WD repeat domain 74
|
|
chr12_-_10282680
|
0.178
|
NM_022570
NM_197947
NM_197948
NM_197949
NM_197950
NM_197954
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chr8_+_68864341
|
0.173
|
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2
|
|
chr7_-_143966380
|
0.171
|
NM_198495
|
CTAGE4
CTAGE8
|
CTAGE family, member 4
CTAGE family, member 8
|
|
chr7_+_143880547
|
0.169
|
NM_198495
|
CTAGE4
CTAGE8
|
CTAGE family, member 4
CTAGE family, member 8
|
|
chr3_-_47957475
|
0.165
|
|
|
|
|
chr10_-_116286661
|
0.164
|
NM_006720
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr2_+_131862419
|
0.162
|
NM_001100623
NM_017958
|
PLEKHB2
|
pleckstrin homology domain containing, family B (evectins) member 2
|
|
chr8_+_22210311
|
0.155
|
|
PIWIL2
|
piwi-like 2 (Drosophila)
|
|
chr5_-_32388687
|
0.152
|
|
ZFR
|
zinc finger RNA binding protein
|
|
chr14_-_22005035
|
0.152
|
NM_005407
|
SALL2
|
sal-like 2 (Drosophila)
|
|
chr3_+_12392993
|
0.142
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr1_-_109935869
|
0.142
|
NM_001205228
|
SORT1
|
sortilin 1
|
|
chr10_-_116418056
|
0.140
|
NM_002313
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr21_-_27512668
|
0.138
|
NM_001136016
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr1_+_178995055
|
0.136
|
NM_014864
|
FAM20B
|
family with sequence similarity 20, member B
|
|
chr10_-_65028825
|
0.134
|
NM_004241
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr10_+_5238756
|
0.129
|
NM_001818
|
AKR1C4
|
aldo-keto reductase family 1, member C4 (chlordecone reductase; 3-alpha hydroxysteroid dehydrogenase, type I; dihydrodiol dehydrogenase 4)
|
|
chr19_-_3985460
|
0.128
|
NM_001961
|
EEF2
|
eukaryotic translation elongation factor 2
|
|
chr1_+_50575465
|
0.126
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr3_-_194188967
|
0.122
|
NM_024524
|
ATP13A3
|
ATPase type 13A3
|
|
chr12_-_10978867
|
0.122
|
NM_023921
|
TAS2R10
|
taste receptor, type 2, member 10
|
|
chr6_+_144612872
|
0.121
|
NM_007124
|
UTRN
|
utrophin
|
|
chr6_+_64231626
|
0.116
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr20_-_47894591
|
0.116
|
NM_021035
|
ZNFX1
|
zinc finger, NFX1-type containing 1
|
|
chrX_+_118108595
|
0.116
|
NM_001031855
NM_024778
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr8_-_17555158
|
0.110
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr7_-_143599163
|
0.110
|
NM_014719
NM_001206938
|
FAM115A
|
family with sequence similarity 115, member A
|
|
chr17_-_73179093
|
0.109
|
NM_001005849
NM_006937
|
SUMO2
|
SMT3 suppressor of mif two 3 homolog 2 (S. cerevisiae)
|
|
chr16_-_53537112
|
0.109
|
NM_001012398
NM_022476
|
AKTIP
|
AKT interacting protein
|
|
chr2_-_160654736
|
0.107
|
NM_001198763
NM_001198764
NM_014880
|
CD302
|
CD302 molecule
|
|
chr9_-_112191105
|
0.105
|
NM_001145371
NM_001145372
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr16_+_70333058
|
0.104
|
NM_001014449
NM_001014451
NM_007242
|
DDX19B
|
DEAD (Asp-Glu-Ala-As) box polypeptide 19B
|
|
chrM_+_1670
|
0.103
|
|
|
|
|
chr7_-_143454771
|
0.102
|
NM_178561
|
CTAGE6P
|
CTAGE family, member 6, pseudogene
|
|
chr9_+_71819926
|
0.102
|
NM_001170415
NM_001170416
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr3_-_123339423
|
0.102
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr12_-_16757944
|
0.100
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr7_-_72936538
|
0.100
|
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B
|
|
chrX_-_153775002
|
0.099
|
|
G6PD
|
glucose-6-phosphate dehydrogenase
|
|
chr6_+_126307575
|
0.097
|
NM_001031712
|
TRMT11
|
tRNA methyltransferase 11 homolog (S. cerevisiae)
|
|
chr1_+_65613211
|
0.095
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr5_-_140013034
|
0.095
|
NM_000591
|
CD14
|
CD14 molecule
|
|
chr18_+_22040592
|
0.093
|
NM_001143828
NM_001160166
NM_021624
|
HRH4
|
histamine receptor H4
|
|
chr6_+_24403144
|
0.092
|
NM_020662
|
MRS2
|
MRS2 magnesium homeostasis factor homolog (S. cerevisiae)
|
|
chr7_-_37956515
|
0.092
|
NM_003014
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr7_+_143268872
|
0.091
|
NM_001008747
|
CTAGE6P
CTAGE15P
|
CTAGE family, member 6, pseudogene
CTAGE family, member 15, pseudogene
|
|
chr12_-_91505215
|
0.091
|
|
LUM
|
lumican
|
|
chr20_-_44455932
|
0.089
|
NM_003279
|
TNNC2
|
troponin C type 2 (fast)
|
|
chr17_-_36956145
|
0.088
|
NM_003559
|
PIP4K2B
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta
|
|
chr2_-_178753464
|
0.088
|
NM_001077196
|
PDE11A
|
phosphodiesterase 11A
|
|
chr9_-_130712866
|
0.088
|
NM_203305
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr5_+_36606667
|
0.087
|
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr3_-_114790221
|
0.086
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr12_-_71551564
|
0.085
|
|
TSPAN8
|
tetraspanin 8
|
|
chr13_+_97928414
|
0.084
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr11_-_62607627
|
0.083
|
NM_018093
|
WDR74
|
WD repeat domain 74
|
|
chr2_-_228582698
|
0.083
|
NM_025243
|
SLC19A3
|
solute carrier family 19, member 3
|
|
chr2_+_201756639
|
0.080
|
NM_001142356
|
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae)
|
|
chr18_-_55289014
|
0.079
|
|
NARS
|
asparaginyl-tRNA synthetase
|
|
chr8_-_103666018
|
0.079
|
NM_001032282
|
KLF10
|
Kruppel-like factor 10
|
|
chr8_+_17396285
|
0.077
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr17_-_38821286
|
0.077
|
NM_152349
|
KRT222
|
keratin 222
|
|
chr6_+_43395285
|
0.076
|
NM_001198934
|
ABCC10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10
|
|
chr5_+_154092461
|
0.076
|
NM_015315
|
LARP1
|
La ribonucleoprotein domain family, member 1
|
|
chr3_+_154797910
|
0.071
|
NM_007288
|
MME
|
membrane metallo-endopeptidase
|
|
chr8_-_72274466
|
0.070
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr19_+_34710327
|
0.070
|
|
LSM14A
|
LSM14A, SCD6 homolog A (S. cerevisiae)
|
|
chr3_+_169684549
|
0.069
|
NM_003262
|
SEC62
|
SEC62 homolog (S. cerevisiae)
|
|
chr14_+_29241989
|
0.067
|
|
C14orf23
|
chromosome 14 open reading frame 23
|
|
chr15_-_83621350
|
0.067
|
NM_004839
NM_199330
|
HOMER2
|
homer homolog 2 (Drosophila)
|
|
chr3_-_11623829
|
0.067
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr2_+_161993465
|
0.066
|
NM_004180
NM_133484
|
TANK
|
TRAF family member-associated NFKB activator
|
|
chr5_-_137848517
|
0.066
|
|
ETF1
|
eukaryotic translation termination factor 1
|
|
chr1_+_87012758
|
0.065
|
NM_012128
|
CLCA4
|
chloride channel accessory 4
|
|
chr9_-_32550818
|
0.064
|
|
TOPORS
|
topoisomerase I binding, arginine/serine-rich, E3 ubiquitin protein ligase
|
|
chr6_-_132032156
|
0.064
|
NM_001145659
|
CTAGE9
|
CTAGE family, member 9
|
|
chr12_-_16758312
|
0.061
|
NM_001243611
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr1_+_7844713
|
0.060
|
NM_016831
|
PER3
|
period homolog 3 (Drosophila)
|
|
chr10_+_112344076
|
0.060
|
|
SMC3
|
structural maintenance of chromosomes 3
|
|
chr19_+_58513428
|
0.059
|
|
LOC100128398
|
uncharacterized LOC100128398
|
|
chr22_+_42229105
|
0.059
|
NM_004599
|
SREBF2
|
sterol regulatory element binding transcription factor 2
|