|
chr1_+_151032866
|
2.856
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr1_+_151032880
|
2.587
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr1_+_151032683
|
2.107
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr18_-_53257033
|
2.101
|
NM_001243227
NM_001243228
|
TCF4
|
transcription factor 4
|
|
chr1_-_26232890
|
2.037
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr12_-_21810720
|
1.801
|
NM_001174097
|
LDHB
|
lactate dehydrogenase B
|
|
chr12_-_21810733
|
1.784
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr12_-_21810754
|
1.780
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr12_-_21810769
|
1.732
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr12_-_21810779
|
1.707
|
NM_002300
|
LDHB
|
lactate dehydrogenase B
|
|
chr1_-_26232643
|
1.538
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr13_-_31040035
|
1.537
|
NM_002128
|
HMGB1
|
high mobility group box 1
|
|
chr14_-_23388380
|
1.469
|
NM_001077351
NM_001077352
NM_018107
|
RBM23
|
RNA binding motif protein 23
|
|
chr12_-_6233684
|
1.469
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr7_-_131241321
|
1.466
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr14_-_23388359
|
1.447
|
|
RBM23
|
RNA binding motif protein 23
|
|
chr1_-_26233367
|
1.446
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr11_+_114167168
|
1.411
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr14_-_23388294
|
1.363
|
|
RBM23
|
RNA binding motif protein 23
|
|
chr1_-_153643456
|
1.311
|
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr13_-_31039966
|
1.295
|
|
HMGB1
|
high mobility group box 1
|
|
chr1_-_153643433
|
1.271
|
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr7_-_44887663
|
1.246
|
|
H2AFV
|
H2A histone family, member V
|
|
chr1_-_153643428
|
1.229
|
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr11_+_114167197
|
1.229
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr1_-_153643474
|
1.220
|
NM_004515
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr18_-_53177999
|
1.209
|
NM_001243231
|
TCF4
|
transcription factor 4
|
|
chr17_-_73150493
|
1.209
|
|
HN1
|
hematological and neurological expressed 1
|
|
chr1_-_153643417
|
1.206
|
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chrX_+_15518899
|
1.150
|
NM_203281
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr14_-_106994001
|
1.127
|
|
IGHV3-48
IGH@
IGHA1
IGHM
|
immunoglobulin heavy variable 3-48
immunoglobulin heavy locus
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant mu
|
|
chr14_-_107131217
|
1.127
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr18_-_52969714
|
1.111
|
NM_001243236
|
TCF4
|
transcription factor 4
|
|
chr3_-_121379757
|
1.090
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr16_-_103557
|
1.072
|
NM_016310
|
POLR3K
|
polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa
|
|
chr1_+_151032150
|
1.066
|
NM_006818
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr11_-_122932806
|
1.066
|
|
HSPA8
|
heat shock 70kDa protein 8
|
|
chr16_+_30194925
|
1.060
|
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr11_-_122932844
|
1.036
|
NM_006597
NM_153201
|
HSPA8
|
heat shock 70kDa protein 8
|
|
chr6_+_80714389
|
1.028
|
|
TTK
|
TTK protein kinase
|
|
chr14_+_24641204
|
1.020
|
NM_001048205
NM_005132
|
REC8
|
REC8 homolog (yeast)
|
|
chr15_-_35087748
|
1.010
|
NM_005159
|
ACTC1
|
actin, alpha, cardiac muscle 1
|
|
chr7_+_99775346
|
1.009
|
|
STAG3
|
stromal antigen 3
|
|
chr1_-_221915393
|
1.000
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr14_-_106926393
|
0.997
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr1_-_221915458
|
0.977
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr10_+_17271276
|
0.975
|
|
VIM
|
vimentin
|
|
chr5_+_177631534
|
0.954
|
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B
|
|
chr3_-_121379701
|
0.943
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr4_+_154387450
|
0.940
|
NM_001131007
NM_015196
|
KIAA0922
|
KIAA0922
|
|
chr10_+_17271277
|
0.934
|
|
VIM
|
vimentin
|
|
chr7_-_27205123
|
0.923
|
|
HOXA9
|
homeobox A9
|
|
chr12_-_6960406
|
0.913
|
NM_031299
|
CDCA3
|
cell division cycle associated 3
|
|
chr3_+_48507621
|
0.905
|
|
TREX1
|
three prime repair exonuclease 1
|
|
chr1_+_101185195
|
0.899
|
NM_001078
NM_001199834
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr14_+_24641084
|
0.898
|
|
REC8
|
REC8 homolog (yeast)
|
|
chr10_+_17271302
|
0.894
|
|
VIM
|
vimentin
|
|
chr5_+_177631533
|
0.886
|
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B
|
|
chr7_-_44887697
|
0.884
|
NM_012412
NM_138635
NM_201436
NM_201516
NM_201517
|
H2AFV
|
H2A histone family, member V
|
|
chr15_-_45670624
|
0.884
|
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase)
|
|
chr7_-_27205148
|
0.884
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr1_-_221915426
|
0.882
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr5_+_177631496
|
0.880
|
NM_004499
NM_031266
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B
|
|
chr2_-_61765394
|
0.879
|
NM_003400
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr16_+_30194730
|
0.866
|
NM_001193333
NM_007074
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr7_-_150329285
|
0.862
|
|
GIMAP6
|
GTPase, IMAP family member 6
|
|
chr10_+_11047253
|
0.859
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr4_-_103266577
|
0.852
|
NM_001135146
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8
|
|
chr8_+_26435590
|
0.850
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr12_+_98909410
|
0.845
|
|
TMPO
|
thymopoietin
|
|
chr8_+_55370494
|
0.845
|
NM_022454
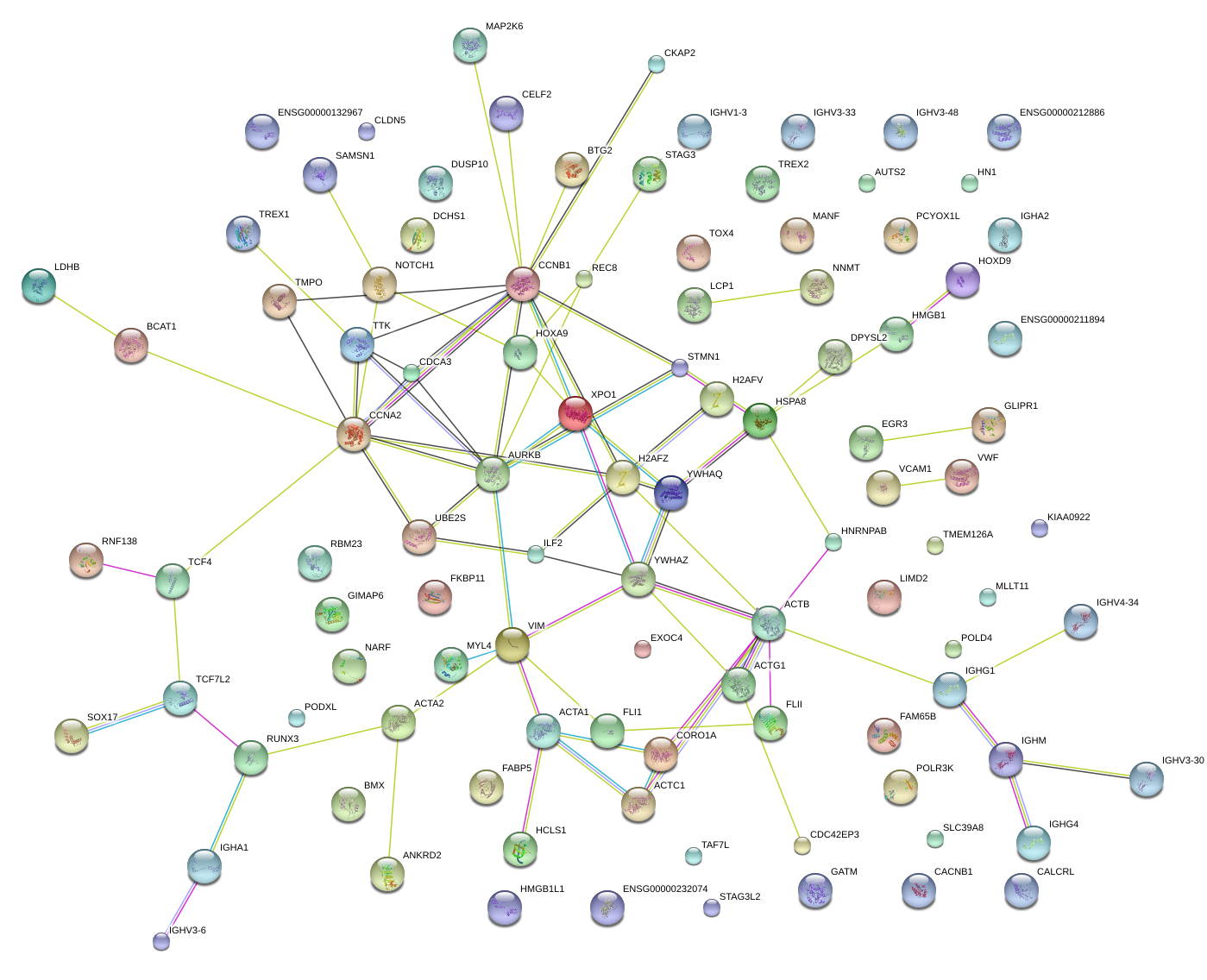
|
SOX17
|
SRY (sex determining region Y)-box 17
|
|
chr17_+_80416530
|
0.844
|
NM_001083608
NM_012336
NM_031968
|
NARF
|
nuclear prelamin A recognition factor
|
|
chr12_+_98909405
|
0.833
|
|
TMPO
|
thymopoietin
|
|
chr21_-_15918635
|
0.832
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr2_-_37898768
|
0.830
|
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr13_+_53029494
|
0.827
|
NM_001098525
NM_018204
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr17_-_8113862
|
0.820
|
NM_004217
|
AURKB
|
aurora kinase B
|
|
chr1_+_203274646
|
0.813
|
NM_006763
|
BTG2
|
BTG family, member 2
|
|
chr17_+_45286713
|
0.800
|
NM_002476
|
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic
|
|
chr7_+_69064316
|
0.793
|
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr11_+_128563772
|
0.787
|
NM_001167681
NM_002017
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr2_-_188312872
|
0.786
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr5_+_68462948
|
0.779
|
|
CCNB1
|
cyclin B1
|
|
chr10_+_11047274
|
0.777
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr17_+_80416606
|
0.769
|
|
NARF
|
nuclear prelamin A recognition factor
|
|
chr2_-_9771101
|
0.767
|
NM_006826
|
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide
|
|
chr10_+_17271956
|
0.761
|
|
VIM
|
vimentin
|
|
chr12_-_25055228
|
0.758
|
NM_001178094
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr11_-_67120943
|
0.753
|
NM_021173
|
POLD4
|
polymerase (DNA-directed), delta 4
|
|
chr13_-_46756295
|
0.753
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr8_+_82192717
|
0.752
|
NM_001444
|
FABP5
|
fatty acid binding protein 5 (psoriasis-associated)
|
|
chr3_+_48507209
|
0.747
|
NM_016381
NM_033629
|
TREX1
|
three prime repair exonuclease 1
|
|
chr12_+_75874512
|
0.744
|
NM_006851
|
GLIPR1
|
GLI pathogenesis-related 1
|
|
chr5_+_68462985
|
0.737
|
|
CCNB1
|
cyclin B1
|
|
chr8_-_101965096
|
0.729
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr4_-_122744782
|
0.728
|
|
CCNA2
|
cyclin A2
|
|
chr19_-_55919324
|
0.720
|
NM_014501
|
UBE2S
|
ubiquitin-conjugating enzyme E2S
|
|
chr3_+_51422716
|
0.711
|
|
MANF
|
mesencephalic astrocyte-derived neurotrophic factor
|
|
chr22_-_19512859
|
0.710
|
NM_001130861
NM_003277
|
CLDN5
|
claudin 5
|
|
chrX_-_15872905
|
0.707
|
|
|
|
|
chr5_+_148737569
|
0.706
|
NM_024028
|
PCYOX1L
|
prenylcysteine oxidase 1 like
|
|
chr10_+_17271859
|
0.704
|
|
VIM
|
vimentin
|
|
chr6_-_24877453
|
0.703
|
NM_015864
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr17_-_61777478
|
0.700
|
NM_030576
|
LIMD2
|
LIM domain containing 2
|
|
chr17_+_67498391
|
0.696
|
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr1_-_25291474
|
0.692
|
NM_001031680
|
RUNX3
|
runt-related transcription factor 3
|
|
chr11_-_6676851
|
0.681
|
NM_003737
|
DCHS1
|
dachsous 1 (Drosophila)
|
|
chr8_-_22550708
|
0.679
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr5_+_102465256
|
0.676
|
NM_015216
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2
|
|
chr9_+_90112754
|
0.670
|
NM_004938
|
DAPK1
|
death-associated protein kinase 1
|
|
chr14_-_91884138
|
0.670
|
|
CCDC88C
|
coiled-coil domain containing 88C
|
|
chr7_+_120590816
|
0.669
|
NM_019071
NM_198267
|
ING3
|
inhibitor of growth family, member 3
|
|
chr10_+_31610063
|
0.667
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr10_+_71078575
|
0.666
|
NM_000188
|
HK1
|
hexokinase 1
|
|
chr2_-_9770999
|
0.665
|
|
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide
|
|
chrX_+_12993224
|
0.665
|
NM_021109
|
TMSB4X
|
thymosin beta 4, X-linked
|
|
chr15_-_45670955
|
0.661
|
NM_001482
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase)
|
|
chr3_+_51422691
|
0.660
|
NM_006010
|
MANF
|
mesencephalic astrocyte-derived neurotrophic factor
|
|
chr7_-_44887567
|
0.656
|
|
H2AFV
|
H2A histone family, member V
|
|
chrX_-_15872921
|
0.654
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr5_+_148737613
|
0.644
|
|
PCYOX1L
|
prenylcysteine oxidase 1 like
|
|
chr10_-_75410778
|
0.643
|
NM_024875
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr1_+_25071759
|
0.642
|
NM_013943
|
CLIC4
|
chloride intracellular channel 4
|
|
chr12_-_21654484
|
0.640
|
NM_002907
NM_032941
|
RECQL
|
RecQ protein-like (DNA helicase Q1-like)
|
|
chr5_+_40680021
|
0.637
|
NM_000958
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr7_-_44887620
|
0.636
|
|
H2AFV
|
H2A histone family, member V
|
|
chr10_+_17271341
|
0.623
|
|
VIM
|
vimentin
|
|
chr2_+_33738941
|
0.623
|
NM_015376
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr1_+_25071906
|
0.623
|
|
CLIC4
|
chloride intracellular channel 4
|
|
chr14_-_106691698
|
0.622
|
|
|
|
|
chr2_-_175869910
|
0.620
|
NM_001822
NM_001025201
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr14_+_85996454
|
0.618
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr1_+_25071967
|
0.618
|
|
CLIC4
|
chloride intracellular channel 4
|
|
chrX_-_15872882
|
0.616
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr16_+_103892
|
0.615
|
|
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12)
|
|
chr2_+_27008881
|
0.612
|
NM_001809
NM_001042426
|
CENPA
|
centromere protein A
|
|
chr12_+_98909331
|
0.611
|
NM_001032283
NM_001032284
NM_003276
|
TMPO
|
thymopoietin
|
|
chr14_+_85996512
|
0.609
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr16_+_103859
|
0.609
|
|
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12)
|
|
chrX_-_15872938
|
0.607
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr2_+_33661392
|
0.604
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr5_-_137071703
|
0.602
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr5_+_112312392
|
0.599
|
NM_001242377
NM_152624
|
DCP2
|
DCP2 decapping enzyme homolog (S. cerevisiae)
|
|
chr6_+_80714321
|
0.599
|
NM_001166691
NM_003318
|
TTK
|
TTK protein kinase
|
|
chr3_+_157154579
|
0.598
|
NM_002852
|
PTX3
|
pentraxin 3, long
|
|
chr4_-_186732208
|
0.596
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr12_-_21654206
|
0.593
|
|
RECQL
|
RecQ protein-like (DNA helicase Q1-like)
|
|
chr7_+_150264457
|
0.592
|
NM_018326
|
GIMAP4
|
GTPase, IMAP family member 4
|
|
chr7_-_120498128
|
0.592
|
NM_012338
|
TSPAN12
|
tetraspanin 12
|
|
chr17_+_1733263
|
0.591
|
NM_002945
|
RPA1
|
replication protein A1, 70kDa
|
|
chr7_+_116660291
|
0.590
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr17_+_1733313
|
0.588
|
|
RPA1
|
replication protein A1, 70kDa
|
|
chr4_-_186732047
|
0.584
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr15_+_85923739
|
0.584
|
NM_006738
NM_007200
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr4_-_40936640
|
0.580
|
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr14_-_106725620
|
0.579
|
|
IGHV4-31
IGHV3-23
IGHA1
IGHG1
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 3-23
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr2_-_158300517
|
0.577
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr7_+_116660263
|
0.572
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr14_+_20937537
|
0.571
|
NM_000270
|
PNP
|
purine nucleoside phosphorylase
|
|
chr1_+_43824598
|
0.570
|
NM_001255
|
CDC20
|
cell division cycle 20 homolog (S. cerevisiae)
|
|
chr13_-_46756326
|
0.565
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr15_-_69113213
|
0.565
|
|
ANP32A
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A
|
|
chr16_+_56817367
|
0.556
|
NM_001242796
|
NUP93
|
nucleoporin 93kDa
|
|
chr1_+_24286300
|
0.555
|
NM_017761
|
PNRC2
|
proline-rich nuclear receptor coactivator 2
|
|
chr18_-_53071225
|
0.549
|
NM_001243232
|
TCF4
|
transcription factor 4
|
|
chr20_+_9494995
|
0.549
|
NM_001199897
NM_012261
|
LAMP5
|
lysosomal-associated membrane protein family, member 5
|
|
chr9_+_126773851
|
0.546
|
NM_004789
|
LHX2
|
LIM homeobox 2
|
|
chr10_+_3109708
|
0.545
|
NM_002627
|
PFKP
|
phosphofructokinase, platelet
|
|
chr2_+_10262694
|
0.539
|
NM_001165931
|
RRM2
|
ribonucleotide reductase M2
|
|
chr6_-_79944397
|
0.537
|
NM_001201362
NM_001201363
NM_004242
NM_138730
|
HMGN3
|
high mobility group nucleosomal binding domain 3
|
|
chr1_+_44115804
|
0.534
|
|
KDM4A
|
lysine (K)-specific demethylase 4A
|
|
chr10_+_124220990
|
0.531
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr19_+_799281
|
0.530
|
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chr7_+_44788175
|
0.527
|
|
ZMIZ2
|
zinc finger, MIZ-type containing 2
|
|
chr7_-_121036355
|
0.526
|
NM_001040020
NM_014888
|
FAM3C
|
family with sequence similarity 3, member C
|
|
chr2_+_10262853
|
0.526
|
NM_001034
|
RRM2
|
ribonucleotide reductase M2
|
|
chr5_-_88178964
|
0.524
|
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr7_-_121036320
|
0.523
|
|
FAM3C
|
family with sequence similarity 3, member C
|
|
chr11_+_75526236
|
0.523
|
|
UVRAG
|
UV radiation resistance associated gene
|
|
chr2_-_61765345
|
0.522
|
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr20_-_56285576
|
0.522
|
NM_199169
NM_199170
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr2_-_61765367
|
0.522
|
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr7_-_99774821
|
0.520
|
NM_152742
|
GPC2
|
glypican 2
|
|
chr2_+_27008912
|
0.517
|
|
CENPA
|
centromere protein A
|
|
chr13_-_41593405
|
0.516
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr1_+_213123853
|
0.512
|
NM_001136474
NM_001136475
NM_024749
|
VASH2
|
vasohibin 2
|
|
chr6_+_12290528
|
0.510
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr14_-_88459469
|
0.507
|
NM_000153
NM_001201401
|
GALC
|
galactosylceramidase
|
|
chr8_+_133787611
|
0.506
|
|
PHF20L1
|
PHD finger protein 20-like 1
|
|
chr20_+_9495297
|
0.505
|
|
LAMP5
|
lysosomal-associated membrane protein family, member 5
|
|
chr15_-_69113089
|
0.504
|
|
ANP32A
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A
|
|
chr15_+_59397276
|
0.504
|
NM_004701
|
CCNB2
|
cyclin B2
|
|
chr10_+_70748476
|
0.503
|
NM_015634
|
KIAA1279
|
KIAA1279
|
|
chr14_-_107218786
|
0.502
|
|
IGHA1
IGHG1
IGHM
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr13_-_99630243
|
0.502
|
NM_001130048
NM_001130050
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr5_+_68462836
|
0.501
|
NM_031966
|
CCNB1
|
cyclin B1
|
|
chr11_+_75526211
|
0.499
|
NM_003369
|
UVRAG
|
UV radiation resistance associated gene
|
|
chr6_-_79944324
|
0.498
|
|
HMGN3
|
high mobility group nucleosomal binding domain 3
|
|
chr6_+_84569394
|
0.497
|
|
CYB5R4
|
cytochrome b5 reductase 4
|
|
chr4_-_122744978
|
0.497
|
NM_001237
|
CCNA2
|
cyclin A2
|