|
chr5_-_39424930
|
24.249
|
|
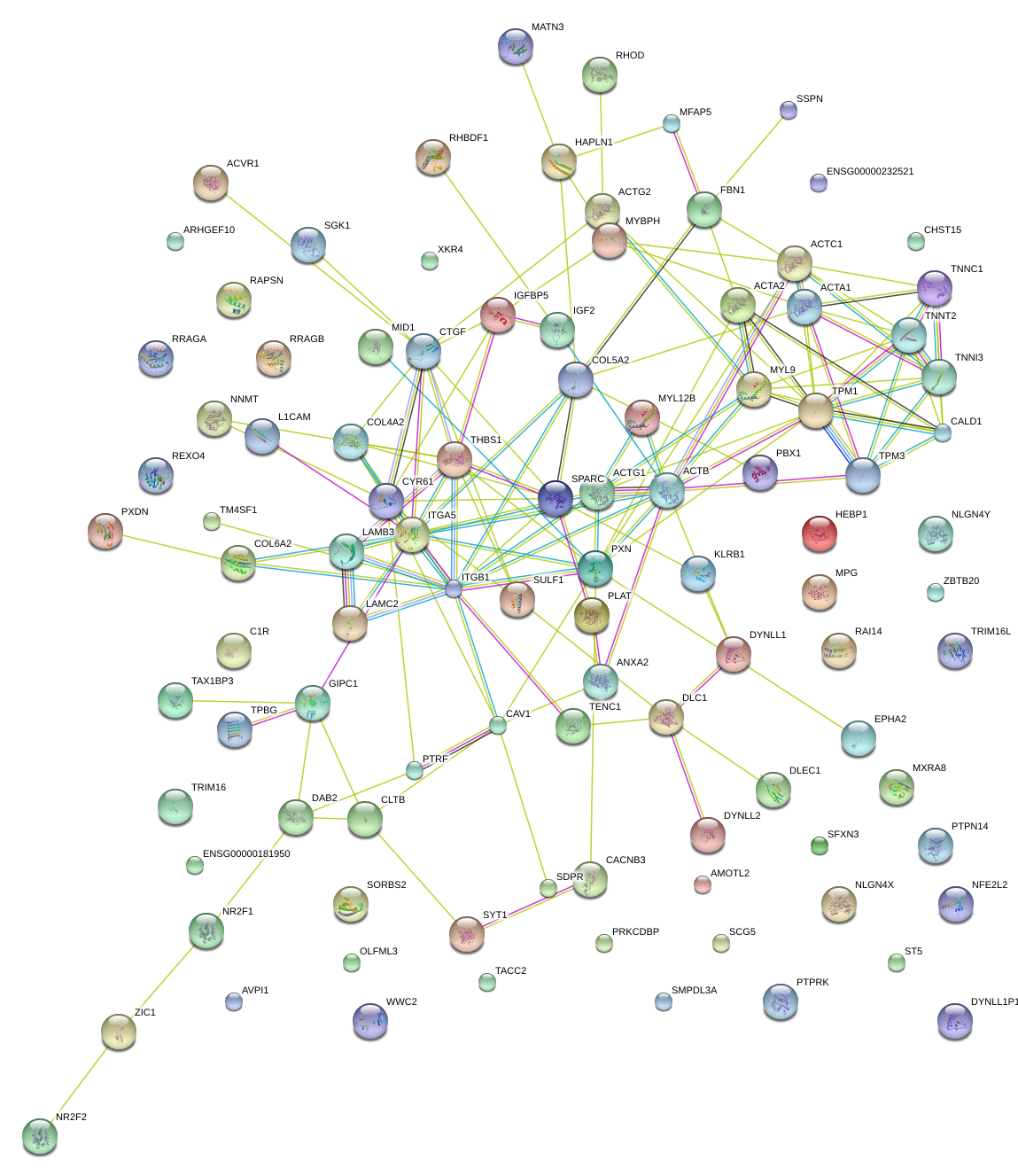
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr6_-_132272311
|
22.068
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr5_-_39425030
|
13.179
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr15_+_39873276
|
11.967
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr6_-_128841432
|
10.814
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr15_+_63334753
|
10.661
|
NM_000366
NM_001018004
NM_001018005
NM_001018006
NM_001018007
NM_001018020
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr15_+_63335086
|
10.289
|
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr11_-_6341708
|
10.213
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr5_-_39425297
|
9.914
|
NM_001244871
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr10_-_90712510
|
9.651
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr11_+_114166534
|
9.389
|
NM_006169
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr2_-_190044330
|
9.150
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr7_+_134464161
|
8.451
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr10_-_125853102
|
8.074
|
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr20_+_35169876
|
8.068
|
NM_006097
NM_181526
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr5_-_151066492
|
7.960
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr20_+_35169897
|
7.703
|
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr7_+_134464417
|
7.350
|
|
CALD1
|
caldesmon 1
|
|
chr12_-_13153200
|
7.254
|
NM_015987
|
HEBP1
|
heme binding protein 1
|
|
chr3_-_149086884
|
7.190
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr1_-_201346804
|
7.159
|
NM_000364
NM_001001430
NM_001001431
NM_001001432
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr1_+_114522029
|
7.095
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chr4_+_184020343
|
6.703
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr1_-_1293907
|
6.689
|
NM_032348
|
MXRA8
|
matrix-remodelling associated 8
|
|
chr8_+_1772095
|
6.625
|
NM_014629
|
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chr1_-_201346783
|
6.581
|
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chrX_-_10588458
|
6.434
|
NM_000381
NM_033289
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr12_-_13153047
|
6.382
|
|
HEBP1
|
heme binding protein 1
|
|
chr2_-_158731622
|
6.229
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr8_-_12973752
|
6.178
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr4_-_186733372
|
6.032
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr5_-_151066445
|
6.006
|
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr5_+_34687663
|
5.963
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr16_-_122573
|
5.962
|
NM_022450
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila)
|
|
chr18_+_8717368
|
5.946
|
NM_015210
|
CCDC165
|
coiled-coil domain containing 165
|
|
chr17_-_40575117
|
5.850
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr11_-_8832881
|
5.736
|
NM_139157
|
ST5
|
suppression of tumorigenicity 5
|
|
chr6_-_134496833
|
5.704
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr3_-_52488030
|
5.650
|
NM_003280
|
TNNC1
|
troponin C type 1 (slow)
|
|
chr10_+_123872440
|
5.640
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr2_-_217540722
|
5.198
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr8_-_42065037
|
5.197
|
|
PLAT
|
plasminogen activator, tissue
|
|
chr10_-_33247178
|
5.082
|
NM_002211
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr3_-_134093235
|
5.007
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr8_-_42065193
|
4.888
|
NM_000930
NM_033011
|
PLAT
|
plasminogen activator, tissue
|
|
chr10_-_33247131
|
4.884
|
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr8_+_70378858
|
4.849
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr10_-_99446899
|
4.753
|
NM_021732
|
AVPI1
|
arginine vasopressin-induced 1
|
|
chr10_+_102792161
|
4.723
|
|
SFXN3
|
sideroflexin 3
|
|
chr6_-_134497069
|
4.661
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr3_-_114343052
|
4.514
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr2_-_192711923
|
4.509
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr17_-_3571819
|
4.416
|
|
TAX1BP3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3
|
|
chr12_-_7245006
|
4.407
|
NM_001733
|
C1R
|
complement component 1, r subcomponent
|
|
chr12_-_8815370
|
4.347
|
NM_003480
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr11_-_8832189
|
4.341
|
NM_213618
|
ST5
|
suppression of tumorigenicity 5
|
|
chr12_-_8815266
|
4.272
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr12_-_7245048
|
4.218
|
|
C1R
|
complement component 1, r subcomponent
|
|
chr15_-_48937795
|
4.091
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr1_+_86046434
|
4.065
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr11_-_2170792
|
3.938
|
NM_001007139
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr12_-_54813002
|
3.927
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr12_+_26348488
|
3.887
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr12_-_8815479
|
3.876
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr2_-_192711651
|
3.782
|
|
SDPR
|
serum deprivation response
|
|
chr7_+_116164838
|
3.677
|
NM_001753
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chrX_-_6146655
|
3.619
|
NM_181332
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr12_-_54812936
|
3.614
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr15_-_60690115
|
3.569
|
NM_001002857
NM_001002858
NM_001136015
NM_004039
|
ANXA2
|
annexin A2
|
|
chr10_+_102792166
|
3.557
|
|
SFXN3
|
sideroflexin 3
|
|
chr6_+_123109970
|
3.526
|
NM_006714
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A
|
|
chr11_-_47470687
|
3.489
|
NM_005055
NM_032645
|
RAPSN
|
receptor-associated protein of the synapse
|
|
chr1_-_209825797
|
3.482
|
NM_000228
|
LAMB3
|
laminin, beta 3
|
|
chr12_-_54813010
|
3.367
|
NM_002205
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr6_+_83073501
|
3.354
|
|
TPBG
|
trophoblast glycoprotein
|
|
chr1_+_183155393
|
3.353
|
|
LAMC2
|
laminin, gamma 2
|
|
chr7_+_134464372
|
3.295
|
|
CALD1
|
caldesmon 1
|
|
chr4_-_186578122
|
3.278
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_-_203144910
|
3.251
|
NM_004997
|
MYBPH
|
myosin binding protein H
|
|
chr17_-_15554809
|
3.244
|
|
TRIM16
|
tripartite motif containing 16
|
|
chr1_-_16482426
|
3.221
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr2_-_20212408
|
3.160
|
|
MATN3
|
matrilin 3
|
|
chr3_+_147127151
|
3.037
|
NM_003412
|
ZIC1
|
Zic family member 1
|
|
chr1_-_209825673
|
3.023
|
NM_001127641
|
LAMB3
|
laminin, beta 3
|
|
chr13_+_111138079
|
3.016
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr12_+_49212253
|
2.973
|
NM_000725
NM_001206915
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr1_-_214724844
|
2.933
|
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr12_+_79258517
|
2.827
|
|
SYT1
|
synaptotagmin I
|
|
chr9_+_19049424
|
2.797
|
|
RRAGA
|
Ras-related GTP binding A
|
|
chrX_-_153141398
|
2.716
|
NM_000425
NM_001143963
NM_024003
|
L1CAM
|
L1 cell adhesion molecule
|
|
chr15_+_96869156
|
2.696
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr12_+_79258588
|
2.684
|
|
SYT1
|
synaptotagmin I
|
|
chr1_+_164528586
|
2.647
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr15_+_32933869
|
2.612
|
NM_001144757
NM_003020
|
SCG5
|
secretogranin V (7B2 protein)
|
|
chr10_+_123748688
|
2.537
|
NM_206861
NM_206862
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr21_+_47545189
|
2.530
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr5_-_175843360
|
2.506
|
|
CLTB
|
clathrin, light chain B
|
|
chr6_+_123110345
|
2.502
|
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A
|
|
chr6_+_83072922
|
2.485
|
NM_006670
|
TPBG
|
trophoblast glycoprotein
|
|
chr12_-_120687956
|
2.473
|
NM_025157
|
PXN
|
paxillin
|
|
chr12_+_53442732
|
2.456
|
NM_015319
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr19_-_14606893
|
2.436
|
|
GIPC1
|
GIPC PDZ domain containing family, member 1
|
|
chr19_-_14606939
|
2.432
|
NM_005716
NM_202467
NM_202468
NM_202469
NM_202470
NM_202494
|
GIPC1
|
GIPC PDZ domain containing family, member 1
|
|
chr5_-_83016850
|
2.373
|
NM_001884
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chr11_+_66824308
|
2.373
|
|
RHOD
|
ras homolog gene family, member D
|
|
chr8_+_56015013
|
2.371
|
NM_052898
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4
|
|
chr11_+_66824285
|
2.364
|
NM_014578
|
RHOD
|
ras homolog gene family, member D
|
|
chr16_+_15528324
|
2.348
|
NM_033201
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr8_-_108510094
|
2.348
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr5_-_55290601
|
2.317
|
NM_001190981
NM_002184
NM_175767
|
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor)
|
|
chr12_+_79258448
|
2.248
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr6_+_134210264
|
2.243
|
|
TCF21
|
transcription factor 21
|
|
chr6_+_134210247
|
2.214
|
NM_198392
NM_003206
|
TCF21
|
transcription factor 21
|
|
chr19_+_45349584
|
2.210
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chrX_-_10851808
|
2.186
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr19_-_51456159
|
2.170
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr7_+_116165089
|
2.130
|
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr6_+_143999058
|
1.996
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr1_-_17307107
|
1.991
|
NM_001135248
NM_002403
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr3_-_127541160
|
1.977
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr18_+_3247774
|
1.972
|
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric
|
|
chr22_+_45072936
|
1.952
|
|
PRR5
|
proline rich 5 (renal)
|
|
chr19_-_44831907
|
1.920
|
|
ZFP112
|
zinc finger protein 112 homolog (mouse)
|
|
chr12_-_56101465
|
1.892
|
|
ITGA7
|
integrin, alpha 7
|
|
chr11_-_14380030
|
1.886
|
NM_001177315
NM_001102669
|
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2
|
|
chr5_+_140868721
|
1.878
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr7_+_116165043
|
1.867
|
NM_001172895
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr21_-_27423423
|
1.826
|
|
|
|
|
chr11_+_2924504
|
1.753
|
|
SLC22A18
|
solute carrier family 22, member 18
|
|
chr14_-_23904849
|
1.753
|
NM_000257
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta
|
|
chr22_+_45072687
|
1.729
|
NM_001017530
NM_001198721
NM_015366
|
PRR5
|
proline rich 5 (renal)
|
|
chr8_-_116681253
|
1.717
|
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr17_+_45286713
|
1.714
|
NM_002476
|
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic
|
|
chr12_-_56101667
|
1.706
|
NM_001144996
NM_002206
|
ITGA7
|
integrin, alpha 7
|
|
chr4_+_169552752
|
1.696
|
NM_001166109
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr17_+_45286427
|
1.694
|
NM_001002841
|
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic
|
|
chr5_+_34684611
|
1.674
|
NM_001145521
NM_001145525
|
RAI14
|
retinoic acid induced 14
|
|
chr15_+_33010174
|
1.663
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr6_+_158733691
|
1.653
|
NM_001007466
NM_020245
|
TULP4
|
tubby like protein 4
|
|
chr16_+_56598960
|
1.646
|
NM_032935
|
MT4
|
metallothionein 4
|
|
chr12_+_79257772
|
1.623
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr7_-_80548322
|
1.612
|
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr1_+_32043745
|
1.612
|
NM_001204415
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr16_-_47495095
|
1.609
|
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1
|
|
chr19_+_36359346
|
1.561
|
NM_001024807
NM_005166
|
APLP1
|
amyloid beta (A4) precursor-like protein 1
|
|
chr9_+_112887780
|
1.550
|
NM_001136562
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chrX_+_107683015
|
1.543
|
NM_000495
NM_033380
|
COL4A5
|
collagen, type IV, alpha 5
|
|
chr5_-_176924552
|
1.539
|
|
PDLIM7
|
PDZ and LIM domain 7 (enigma)
|
|
chr1_-_227505796
|
1.538
|
NM_003607
NM_014826
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like)
|
|
chr12_+_27677044
|
1.522
|
NM_001198915
NM_001198916
NM_003622
NM_177444
|
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1)
|
|
chr9_+_34990715
|
1.518
|
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr5_+_140749830
|
1.501
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr12_-_80084742
|
1.499
|
NM_002583
|
PAWR
|
PRKC, apoptosis, WT1, regulator
|
|
chr17_-_48278874
|
1.486
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr6_+_30850693
|
1.484
|
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr6_-_3157696
|
1.433
|
NM_001069
|
TUBB2A
|
tubulin, beta 2A class IIa
|
|
chr7_-_150946031
|
1.413
|
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr4_+_148538564
|
1.406
|
|
TMEM184C
|
transmembrane protein 184C
|
|
chr19_+_50936159
|
1.400
|
NM_004533
|
MYBPC2
|
myosin binding protein C, fast type
|
|
chr3_+_158787040
|
1.390
|
NM_001197113
NM_001197114
NM_001042705
NM_001042706
NM_001197100
|
IQCJ-SCHIP1
IQCJ
|
IQCJ-SCHIP1 readthrough
IQ motif containing J
|
|
chr11_+_7506599
|
1.387
|
NM_198474
|
OLFML1
|
olfactomedin-like 1
|
|
chr17_-_48278987
|
1.379
|
NM_000088
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr2_+_152214105
|
1.370
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr7_+_100464949
|
1.366
|
NM_003302
|
TRIP6
|
thyroid hormone receptor interactor 6
|
|
chr18_+_3247825
|
1.363
|
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric
|
|
chr1_-_201438150
|
1.338
|
NM_012396
|
PHLDA3
|
pleckstrin homology-like domain, family A, member 3
|
|
chr1_+_145524989
|
1.317
|
NM_003637
|
ITGA10
|
integrin, alpha 10
|
|
chr14_-_74180341
|
1.312
|
NM_006029
|
PNMA1
|
paraneoplastic antigen MA1
|
|
chrY_+_16634487
|
1.309
|
NM_001206850
|
NLGN4Y
|
neuroligin 4, Y-linked
|
|
chr19_-_51504799
|
1.308
|
NM_007196
NM_144505
NM_144506
NM_144507
|
KLK8
|
kallikrein-related peptidase 8
|
|
chr1_-_150780705
|
1.293
|
NM_000396
|
CTSK
|
cathepsin K
|
|
chr1_+_19923457
|
1.290
|
NM_001204088
NM_001204089
NM_001032363
NM_001204082
NM_001204083
|
C1orf151-NBL1
MINOS1
|
C1orf151-NBL1 readthrough
mitochondrial inner membrane organizing system 1
|
|
chr1_+_183155361
|
1.278
|
|
LAMC2
|
laminin, gamma 2
|
|
chr11_+_19372270
|
1.278
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr4_-_186577858
|
1.277
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_+_148538508
|
1.265
|
NM_018241
|
TMEM184C
|
transmembrane protein 184C
|
|
chr16_+_22524843
|
1.253
|
NM_001135865
|
LOC100132247
NPIPL3
|
nuclear pore complex interacting protein related gene
nuclear pore complex interacting protein-like 3
|
|
chr17_+_37856253
|
1.248
|
NM_004448
|
ERBB2
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian)
|
|
chr19_-_49371710
|
1.237
|
NM_001161354
NM_020904
|
PLEKHA4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4
|
|
chr12_+_119616594
|
1.234
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr15_-_42749710
|
1.213
|
NM_022473
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr1_-_149908759
|
1.203
|
NM_001145862
|
MTMR11
|
myotubularin related protein 11
|
|
chr6_+_52930160
|
1.197
|
|
FBXO9
|
F-box protein 9
|
|
chr16_-_21436657
|
1.191
|
NM_130464
|
NPIPL3
|
nuclear pore complex interacting protein-like 3
|
|
chr10_+_71812536
|
1.159
|
|
H2AFY2
|
H2A histone family, member Y2
|
|
chr18_+_3247527
|
1.152
|
NM_006471
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric
|
|
chr1_+_218518675
|
1.145
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr18_+_3247815
|
1.129
|
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric
|
|
chr3_+_169940246
|
1.119
|
|
PRKCI
|
protein kinase C, iota
|
|
chr17_+_4901199
|
1.116
|
NM_006612
|
KIF1C
|
kinesin family member 1C
|
|
chr12_-_106533810
|
1.111
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr1_-_149908721
|
1.093
|
|
MTMR11
|
myotubularin related protein 11
|
|
chr3_+_136537860
|
1.085
|
NM_001097599
NM_001097600
NM_025246
|
TMEM22
|
transmembrane protein 22
|
|
chr3_+_169940214
|
1.085
|
NM_002740
|
PRKCI
|
protein kinase C, iota
|
|
chr11_-_6643809
|
1.067
|
|
DCHS1
|
dachsous 1 (Drosophila)
|
|
chr1_+_144614958
|
1.061
|
NM_001037675
|
NBPF9
NBPF8
|
neuroblastoma breakpoint family, member 9
neuroblastoma breakpoint family, member 8
|
|
chr3_-_120169493
|
1.057
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr14_+_76044939
|
1.052
|
NM_017791
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2
|
|
chrX_+_48368063
|
1.052
|
NM_203474
NM_203475
|
PORCN
|
porcupine homolog (Drosophila)
|
|
chr9_+_125137600
|
1.045
|
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|