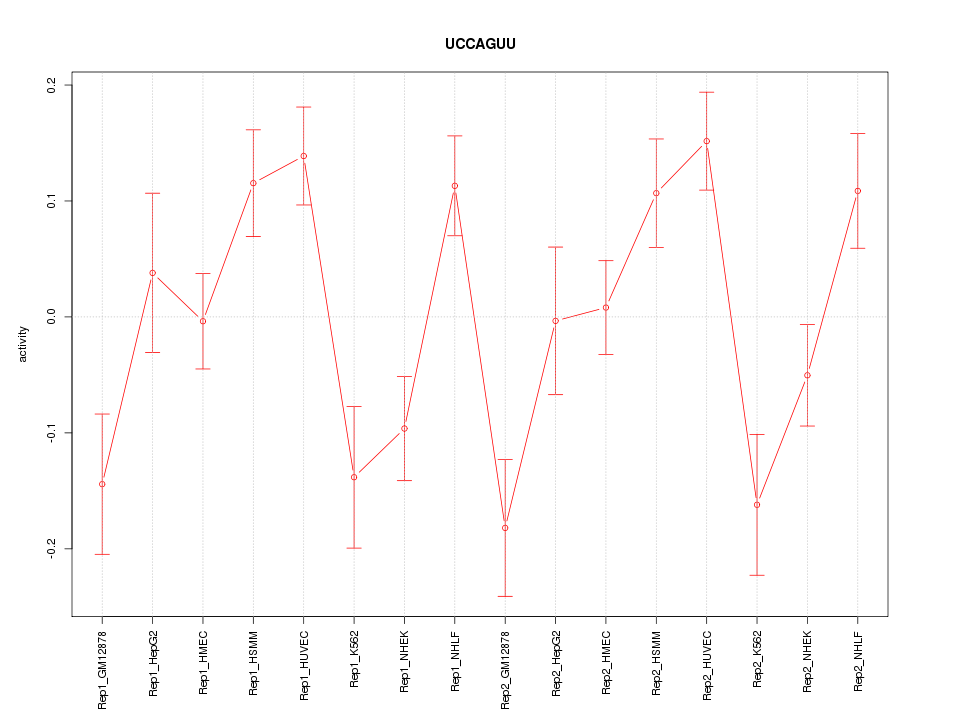
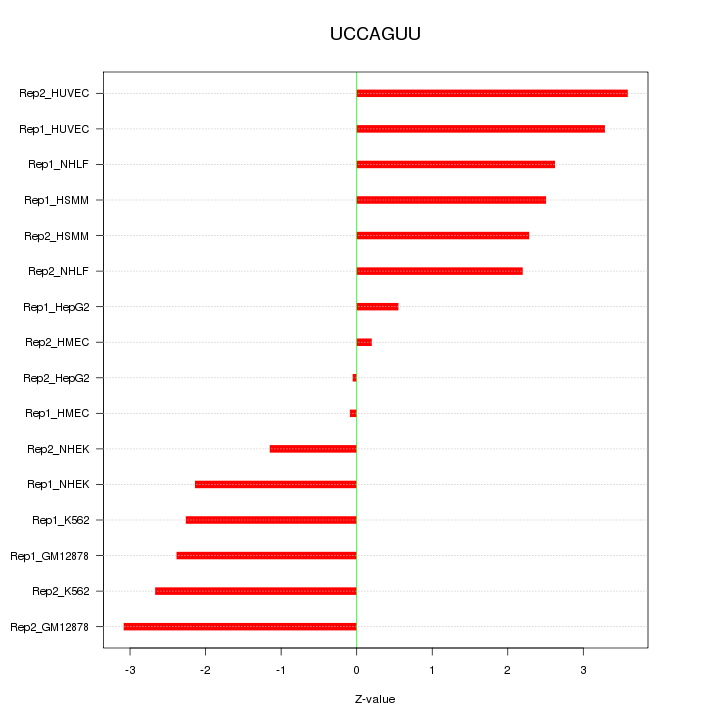
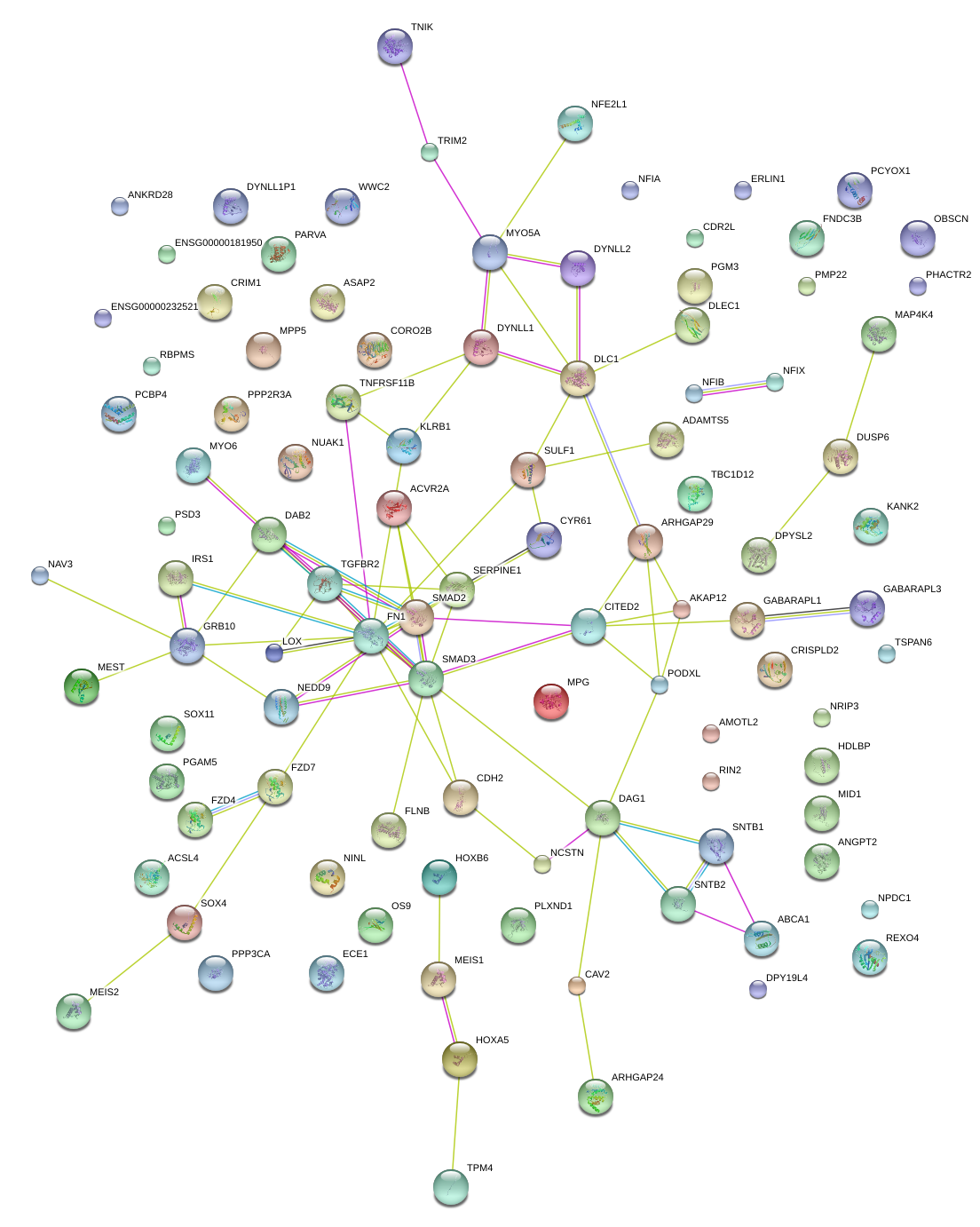
|
chr6_+_151646634
|
8.096
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr6_+_151561094
|
7.775
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr5_-_39425297
|
7.512
|
NM_001244871
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr12_-_106533810
|
6.946
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr7_+_130126035
|
6.591
|
NM_177524
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr5_-_121412805
|
6.424
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr1_+_86046434
|
6.237
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr8_-_12973752
|
6.168
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr8_-_12990731
|
6.051
|
NM_006094
|
DLC1
|
deleted in liver cancer 1
|
|
chr7_+_130131172
|
5.710
|
NM_177525
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr7_+_130131921
|
5.487
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr1_-_94703252
|
5.335
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chrX_-_99891690
|
4.921
|
NM_003270
|
TSPAN6
|
tetraspanin 6
|
|
chr12_+_78225068
|
4.781
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr5_-_121414011
|
4.736
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr7_+_100770378
|
4.459
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr2_+_9346876
|
4.345
|
NM_001135191
NM_003887
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr7_-_131241321
|
4.078
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr3_+_30647901
|
3.491
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr12_-_89746244
|
3.367
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr8_+_26435339
|
3.296
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr18_-_25757351
|
3.296
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr8_-_119964059
|
3.234
|
NM_002546
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b
|
|
chr17_-_46682333
|
3.152
|
NM_018952
|
HOXB6
|
homeobox B6
|
|
chr3_-_129325175
|
3.152
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr8_+_70405027
|
3.133
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr8_+_70378858
|
3.132
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr17_-_15165873
|
3.118
|
NM_153321
|
PMP22
|
peripheral myelin protein 22
|
|
chr8_+_26371461
|
3.023
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr21_-_28339405
|
3.006
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr3_-_134093235
|
2.970
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr17_-_15168590
|
2.945
|
NM_000304
|
PMP22
|
peripheral myelin protein 22
|
|
chr11_-_86666211
|
2.860
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr17_-_15164043
|
2.763
|
NM_153322
|
PMP22
|
peripheral myelin protein 22
|
|
chr2_+_36583369
|
2.763
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr6_-_11382501
|
2.720
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr2_-_227663454
|
2.680
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr9_-_14398981
|
2.620
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr11_+_12399024
|
2.436
|
NM_018222
|
PARVA
|
parvin, alpha
|
|
chr16_+_69220940
|
2.435
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr7_-_27183225
|
2.379
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr6_+_76458888
|
2.351
|
NM_004999
|
MYO6
|
myosin VI
|
|
chr9_-_14314036
|
2.220
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr8_-_18871163
|
2.185
|
NM_015310
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr7_+_116140097
|
2.071
|
NM_001206748
|
CAV2
|
caveolin 2
|
|
chr15_+_67458492
|
2.056
|
NM_001145104
|
SMAD3
|
SMAD family member 3
|
|
chr9_-_107690526
|
1.953
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr7_+_116139619
|
1.894
|
NM_001206747
NM_001233
NM_198212
|
CAV2
|
caveolin 2
|
|
chr20_+_19867075
|
1.871
|
NM_001242581
|
RIN2
|
Ras and Rab interactor 2
|
|
chr6_+_143929316
|
1.867
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr2_-_216300770
|
1.858
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr4_-_102268626
|
1.845
|
NM_000944
NM_001130691
NM_001130692
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr6_+_143999058
|
1.843
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr3_-_51996855
|
1.841
|
NM_033010
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr8_-_18666295
|
1.840
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr12_+_10365415
|
1.810
|
NM_031412
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1
|
|
chr10_-_101945787
|
1.755
|
NM_001100626
|
ERLIN1
|
ER lipid raft associated 1
|
|
chr2_+_66662516
|
1.751
|
NM_002398
|
MEIS1
|
Meis homeobox 1
|
|
chr2_+_102314162
|
1.744
|
NM_001242560
NM_004834
NM_145686
NM_001242559
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr1_-_21616648
|
1.723
|
NM_001113349
|
ECE1
|
endothelin converting enzyme 1
|
|
chr20_+_19870209
|
1.720
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr15_+_67358194
|
1.684
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr2_+_5832778
|
1.641
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr10_-_101945686
|
1.639
|
NM_006459
|
ERLIN1
|
ER lipid raft associated 1
|
|
chr15_+_68924326
|
1.581
|
NM_001190457
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr11_-_9025531
|
1.562
|
NM_020645
|
NRIP3
|
nuclear receptor interacting protein 3
|
|
chr3_-_15900928
|
1.547
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr15_+_68908878
|
1.535
|
NM_001190456
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chrX_-_10588458
|
1.518
|
NM_000381
NM_033289
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr3_-_15798057
|
1.499
|
NM_001195099
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr8_-_121824287
|
1.494
|
NM_021021
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chr4_+_86851425
|
1.492
|
NM_031305
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr1_-_21605984
|
1.491
|
NM_001113347
|
ECE1
|
endothelin converting enzyme 1
|
|
chr4_+_86699846
|
1.477
|
NM_001042669
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr19_-_11308184
|
1.464
|
NM_001136191
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr6_-_139695349
|
1.461
|
NM_001168389
NM_001168388
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr3_+_57994087
|
1.460
|
NM_001164317
NM_001164318
NM_001164319
NM_001457
|
FLNB
|
filamin B, beta
|
|
chr1_+_160313061
|
1.449
|
NM_015331
|
NCSTN
|
nicastrin
|
|
chrX_-_10544852
|
1.448
|
NM_001193277
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr15_+_67418053
|
1.419
|
NM_001145102
|
SMAD3
|
SMAD family member 3
|
|
chr4_+_86396251
|
1.408
|
NM_001025616
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr7_-_50800018
|
1.400
|
NM_001001549
NM_005311
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr2_+_148602569
|
1.386
|
NM_001616
|
ACVR2A
|
activin A receptor, type IIA
|
|
chr6_-_11232901
|
1.381
|
NM_006403
NM_182966
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr17_+_72983723
|
1.375
|
NM_014603
|
CDR2L
|
cerebellar degeneration-related protein 2-like
|
|
chr6_-_139695784
|
1.370
|
NM_006079
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr20_-_25566137
|
1.369
|
NM_025176
|
NINL
|
ninein-like
|
|
chr8_+_30241943
|
1.366
|
NM_001008710
NM_001008711
NM_001008712
NM_006867
|
RBPMS
|
RNA binding protein with multiple splicing
|
|
chr9_-_139940595
|
1.365
|
NM_015392
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chrX_-_108976509
|
1.363
|
NM_004458
NM_022977
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr15_+_67430358
|
1.362
|
NM_001145103
|
SMAD3
|
SMAD family member 3
|
|
chr16_+_84853585
|
1.360
|
NM_031476
|
CRISPLD2
|
cysteine-rich secretory protein LCCL domain containing 2
|
|
chr4_+_154074269
|
1.352
|
NM_001130067
|
TRIM2
|
tripartite motif containing 2
|
|
chr8_-_6420454
|
1.347
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr7_-_50861114
|
1.345
|
NM_001001555
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chrX_-_10851808
|
1.341
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chrX_-_10645726
|
1.333
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr1_-_21671905
|
1.333
|
NM_001113348
|
ECE1
|
endothelin converting enzyme 1
|
|
chr1_-_21616888
|
1.298
|
NM_001397
|
ECE1
|
endothelin converting enzyme 1
|
|
chr15_-_52821004
|
1.264
|
NM_000259
NM_001142495
|
MYO5A
|
myosin VA (heavy chain 12, myoxin)
|
|
chr4_+_184020343
|
1.259
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr19_-_11305186
|
1.255
|
NM_015493
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr19_+_16178170
|
1.235
|
NM_001145160
|
TPM4
|
tropomyosin 4
|
|
chr12_+_58087737
|
1.229
|
NM_001017956
NM_001017957
NM_001017958
NM_006812
|
OS9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin
|
|
chr3_+_49506079
|
1.199
|
NM_001177643
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chr6_-_83903632
|
1.164
|
NM_001199917
|
PGM3
|
phosphoglucomutase 3
|
|
chr8_+_95732101
|
1.157
|
NM_181787
|
DPY19L4
|
dpy-19-like 4 (C. elegans)
|
|
chr19_+_16187047
|
1.139
|
NM_003290
|
TPM4
|
tropomyosin 4
|
|
chr17_+_46125675
|
1.126
|
NM_003204
|
NFE2L1
|
nuclear factor (erythroid-derived 2)-like 1
|
|
chr3_+_135684463
|
1.114
|
NM_001190447
NM_002718
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr10_+_96162185
|
1.102
|
NM_015188
|
TBC1D12
|
TBC1 domain family, member 12
|
|
chr3_+_171757413
|
1.101
|
NM_022763
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr2_-_242255114
|
1.099
|
NM_005336
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr14_+_67707825
|
1.093
|
NM_022474
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5)
|
|
chr4_+_154125589
|
1.092
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr2_+_202899309
|
1.088
|
NM_003507
|
FZD7
|
frizzled family receptor 7
|
|
chr7_-_50772997
|
1.082
|
NM_001001550
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr2_+_70485204
|
1.071
|
NM_016297
|
PCYOX1
|
prenylcysteine oxidase 1
|
|
chrX_+_48432740
|
1.060
|
NM_006743
|
RBM3
|
RNA binding motif (RNP1, RRM) protein 3
|
|
chr3_+_135741576
|
1.060
|
NM_181897
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr21_-_39870303
|
1.039
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr3_-_52001388
|
1.022
|
NM_020418
NM_001174100
NM_033008
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr1_+_89149836
|
1.019
|
NM_006256
|
PKN2
|
protein kinase N2
|
|
chr13_-_72441312
|
1.002
|
NM_004392
NM_080759
NM_080760
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr15_+_99192696
|
0.972
|
NM_000875
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr9_+_112542496
|
0.964
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr11_+_576482
|
0.956
|
NM_020901
|
PHRF1
|
PHD and ring finger domains 1
|
|
chr12_-_120703549
|
0.948
|
NM_001080855
NM_001243756
NM_002859
|
PXN
|
paxillin
|
|
chr6_+_158733691
|
0.900
|
NM_001007466
NM_020245
|
TULP4
|
tubby like protein 4
|
|
chr12_+_52345437
|
0.888
|
NM_004302
NM_020328
|
ACVR1B
|
activin A receptor, type IB
|
|
chr11_+_61447816
|
0.870
|
NM_006133
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chr7_+_872137
|
0.861
|
NM_001130965
|
SUN1
|
Sad1 and UNC84 domain containing 1
|
|
chr2_-_106810781
|
0.853
|
NM_025076
|
UXS1
|
UDP-glucuronate decarboxylase 1
|
|
chr1_-_208417635
|
0.850
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr10_-_118032717
|
0.847
|
NM_001145453
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr19_+_34745452
|
0.824
|
NM_014686
|
KIAA0355
|
KIAA0355
|
|
chrX_+_73641084
|
0.812
|
NM_006517
|
SLC16A2
|
solute carrier family 16, member 2 (monocarboxylic acid transporter 8)
|
|
chr3_+_49507470
|
0.808
|
NM_001165928
NM_001177634
NM_001177635
NM_001177636
NM_001177637
NM_001177638
NM_001177639
NM_001177640
NM_001177641
NM_001177642
NM_001177644
NM_004393
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chr19_+_18794391
|
0.804
|
NM_001098482
NM_015321
|
CRTC1
|
CREB regulated transcription coactivator 1
|
|
chr5_+_86564727
|
0.802
|
NM_022650
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr16_-_49860917
|
0.797
|
NM_015069
|
ZNF423
|
zinc finger protein 423
|
|
chr7_+_94285620
|
0.796
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr9_+_116226006
|
0.796
|
NM_017790
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr11_-_118966175
|
0.776
|
NM_002105
|
H2AFX
|
H2A histone family, member X
|
|
chr5_-_179233772
|
0.767
|
NM_014275
|
MGAT4B
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B
|
|
chr10_-_118032975
|
0.760
|
NM_005264
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr10_-_118031542
|
0.759
|
NM_145793
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr11_+_9406123
|
0.758
|
NM_006391
|
IPO7
|
importin 7
|
|
chr12_+_13043955
|
0.753
|
NM_003979
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr14_-_75179739
|
0.752
|
NM_001039479
|
KIAA0317
|
KIAA0317
|
|
chr1_+_186798031
|
0.748
|
NM_024420
|
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr4_-_53522756
|
0.732
|
NM_001134223
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr5_-_77944341
|
0.717
|
NM_005779
|
LHFPL2
|
lipoma HMGIC fusion partner-like 2
|
|
chr5_+_86564044
|
0.704
|
NM_002890
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr18_+_33234532
|
0.699
|
NM_020474
|
GALNT1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1)
|
|
chr4_+_78078332
|
0.694
|
NM_004354
|
CCNG2
|
cyclin G2
|
|
chr7_+_94285676
|
0.690
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr11_-_82611358
|
0.688
|
NM_005040
NM_199418
|
PRCP
|
prolylcarboxypeptidase (angiotensinase C)
|
|
chr13_-_30424795
|
0.688
|
NM_007106
|
UBL3
|
ubiquitin-like 3
|
|
chr4_+_72204769
|
0.686
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr6_-_116575170
|
0.678
|
NM_021648
|
TSPYL4
|
TSPY-like 4
|
|
chr12_+_72233486
|
0.666
|
NM_001146213
NM_001146214
NM_022771
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr1_+_218518675
|
0.662
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr18_-_812268
|
0.657
|
NM_005433
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1
|
|
chr15_+_63569748
|
0.653
|
NM_001145646
NM_031301
|
APH1B
|
anterior pharynx defective 1 homolog B (C. elegans)
|
|
chr12_-_112037360
|
0.648
|
NM_002973
|
ATXN2
|
ataxin 2
|
|
chr21_-_40033617
|
0.630
|
NM_001243432
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr4_+_81187741
|
0.628
|
NM_004464
NM_033143
|
FGF5
|
fibroblast growth factor 5
|
|
chr3_+_23987514
|
0.616
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr17_-_1090562
|
0.616
|
NM_001159746
|
ABR
|
active BCR-related gene
|
|
chr17_-_1359410
|
0.597
|
NM_005206
NM_016823
|
CRK
|
v-crk sarcoma virus CT10 oncogene homolog (avian)
|
|
chr15_+_68871285
|
0.583
|
NM_006091
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr3_+_69134057
|
0.583
|
NM_006407
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5
|
|
chr13_+_73633097
|
0.579
|
NM_001730
|
KLF5
|
Kruppel-like factor 5 (intestinal)
|
|
chr12_+_52347163
|
0.574
|
NM_020327
|
ACVR1B
|
activin A receptor, type IB
|
|
chr5_-_179229874
|
0.561
|
NM_054013
|
MGAT4B
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B
|
|
chr6_-_111804413
|
0.558
|
NM_002912
|
REV3L
|
REV3-like, catalytic subunit of DNA polymerase zeta (yeast)
|
|
chrX_-_77041678
|
0.552
|
NM_000489
NM_138270
|
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked
|
|
chr17_-_58603492
|
0.550
|
NM_006380
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2
|
|
chrX_-_83442906
|
0.538
|
NM_014496
|
RPS6KA6
|
ribosomal protein S6 kinase, 90kDa, polypeptide 6
|
|
chr11_-_104035023
|
0.533
|
NM_025208
NM_033135
|
PDGFD
|
platelet derived growth factor D
|
|
chr13_-_29069215
|
0.524
|
NM_001159920
NM_001160030
NM_001160031
NM_002019
|
FLT1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor)
|
|
chr12_-_120687956
|
0.519
|
NM_025157
|
PXN
|
paxillin
|
|
chr19_-_58662144
|
0.516
|
NM_024620
|
ZNF329
|
zinc finger protein 329
|
|
chr6_-_83902921
|
0.516
|
NM_001199918
NM_001199919
NM_015599
|
PGM3
|
phosphoglucomutase 3
|
|
chr17_-_1012257
|
0.513
|
NM_001092
|
ABR
|
active BCR-related gene
|
|
chr3_+_179370779
|
0.511
|
NM_003940
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3)
|
|
chr11_-_8985955
|
0.508
|
NM_020644
|
TMEM9B
|
TMEM9 domain family, member B
|
|
chr4_-_156298094
|
0.502
|
NM_001039580
|
MAP9
|
microtubule-associated protein 9
|
|
chr17_-_40307015
|
0.501
|
NM_001252039
NM_004583
NM_201434
|
RAB5C
|
RAB5C, member RAS oncogene family
|
|
chr17_-_5389480
|
0.494
|
NM_016041
|
DERL2
|
Der1-like domain family, member 2
|
|
chr18_+_3247527
|
0.491
|
NM_006471
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric
|
|
chr3_-_11761946
|
0.487
|
NM_014667
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr19_+_1269244
|
0.486
|
NM_001280
|
CIRBP
|
cold inducible RNA binding protein
|
|
chr20_-_4795768
|
0.484
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr8_-_17658385
|
0.480
|
NM_001001924
NM_001001925
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr3_+_171758289
|
0.469
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr12_+_49760679
|
0.463
|
NM_023071
|
SPATS2
|
spermatogenesis associated, serine-rich 2
|
|
chr8_-_17555158
|
0.458
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr12_-_25102281
|
0.450
|
NM_001178091
NM_001178092
NM_005504
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|