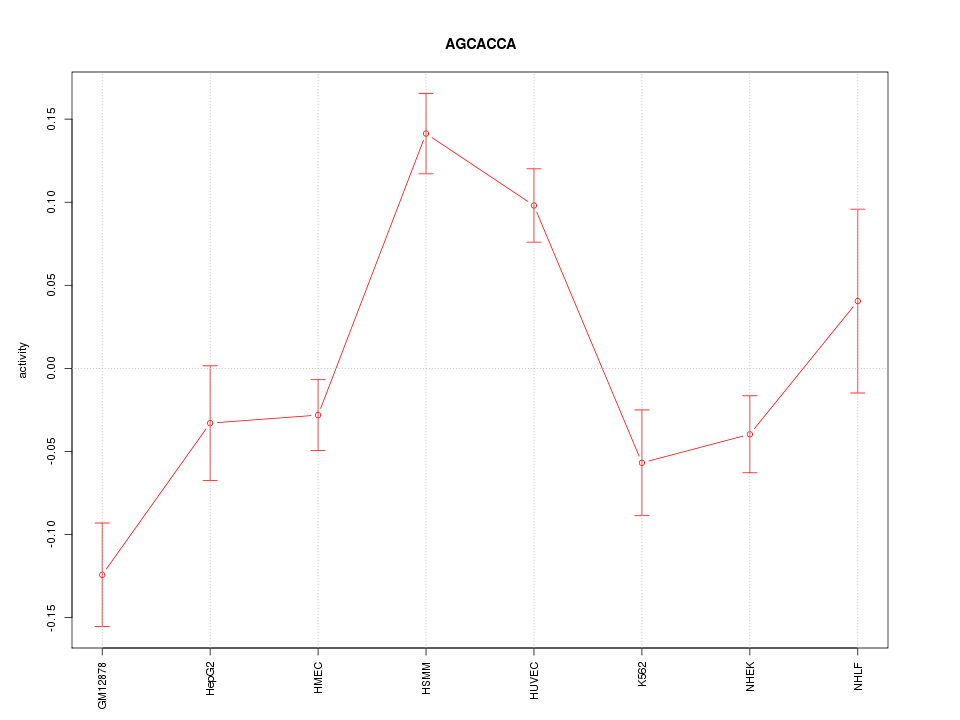
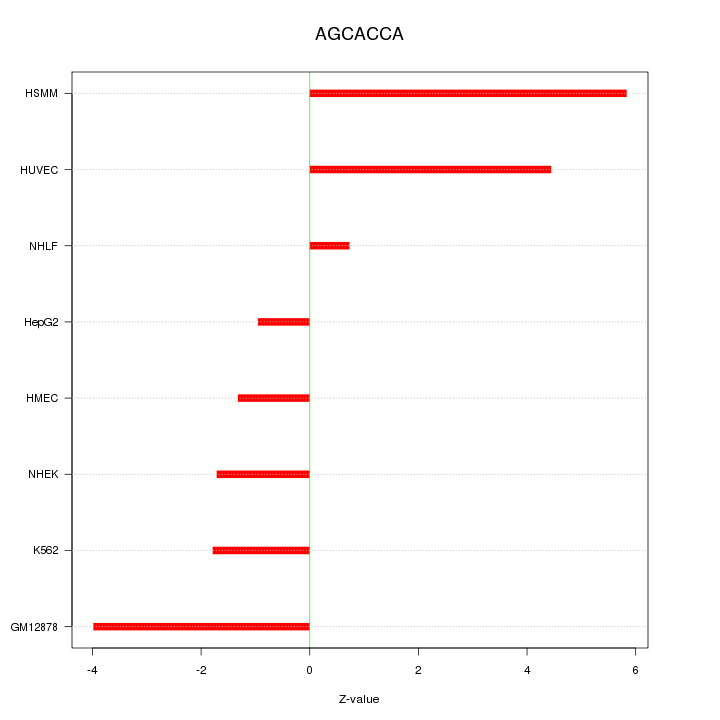
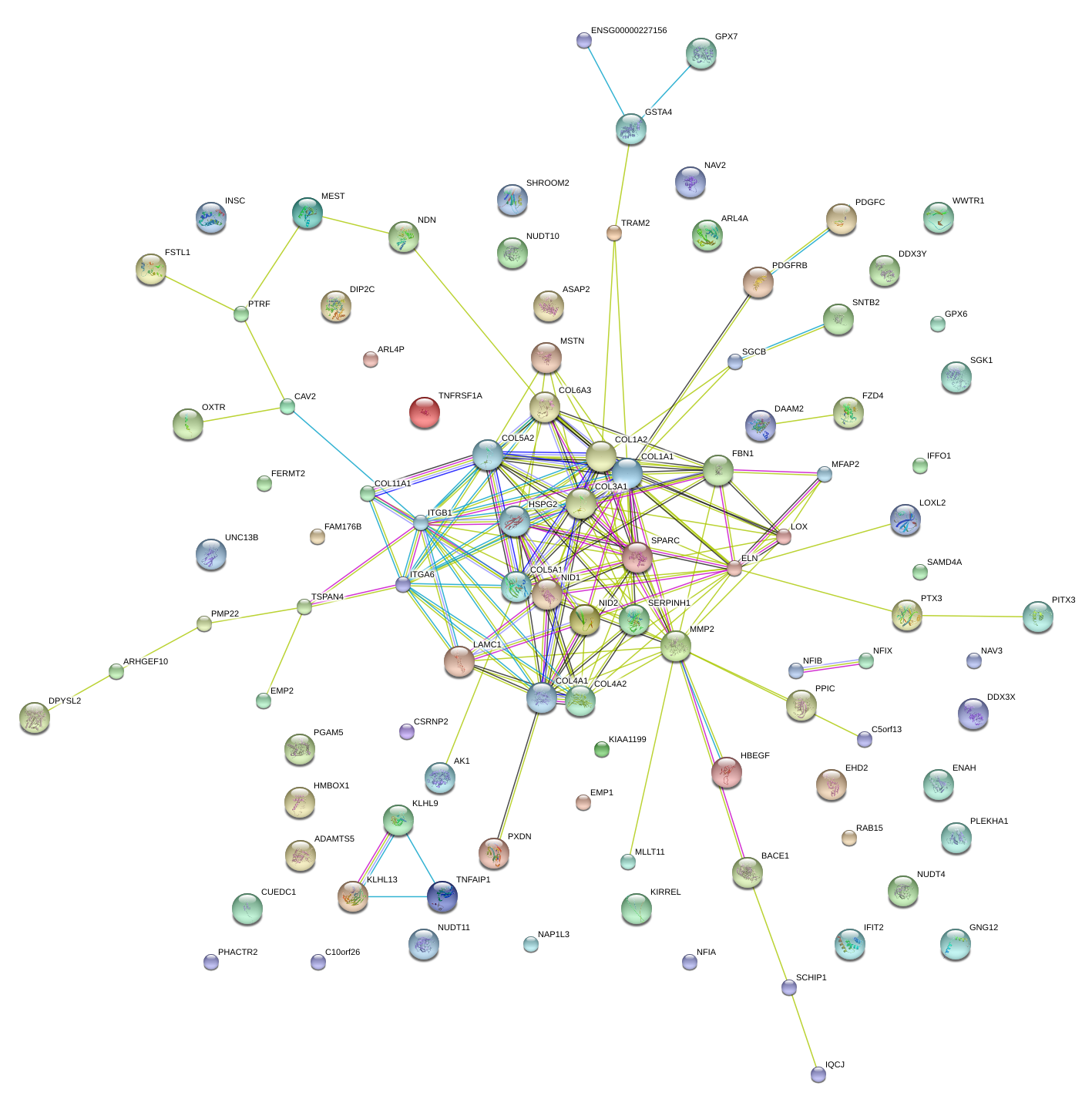
|
chr2_-_190044330
|
7.006
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr3_-_120169493
|
6.592
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr7_+_94023872
|
6.580
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr5_-_151066492
|
6.263
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr3_+_157154579
|
6.215
|
NM_002852
|
PTX3
|
pentraxin 3, long
|
|
chr2_-_1748285
|
6.050
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr13_-_110959441
|
5.809
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr2_+_189839078
|
5.681
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr5_-_121412805
|
5.564
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr9_+_137533643
|
5.514
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr16_+_55515468
|
5.433
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr14_-_52535945
|
5.036
|
NM_007361
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr15_-_48937795
|
5.024
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr12_+_13349601
|
4.847
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr16_+_55512801
|
4.836
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr3_+_159557649
|
4.665
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr3_+_158991035
|
4.630
|
NM_001197107
NM_001197108
NM_014575
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr1_-_68299047
|
4.520
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr15_+_81071708
|
4.397
|
NM_018689
|
KIAA1199
|
KIAA1199
|
|
chr13_+_110959592
|
4.352
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr7_+_130126035
|
4.334
|
NM_177524
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr12_+_78225068
|
4.275
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr1_-_103574051
|
4.252
|
NM_001190709
NM_001854
NM_080629
NM_080630
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr5_-_121414011
|
4.042
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr21_-_28339405
|
3.952
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr7_+_130131172
|
3.815
|
NM_177525
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr7_+_130131921
|
3.603
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr17_-_15165873
|
3.586
|
NM_153321
|
PMP22
|
peripheral myelin protein 22
|
|
chr5_-_122372413
|
3.493
|
NM_000943
|
PPIC
|
peptidylprolyl isomerase C (cyclophilin C)
|
|
chr17_-_15168590
|
3.387
|
NM_000304
|
PMP22
|
peripheral myelin protein 22
|
|
chr4_-_157892545
|
3.367
|
NM_016205
|
PDGFC
|
platelet derived growth factor C
|
|
chr7_+_116140097
|
3.339
|
NM_001206748
|
CAV2
|
caveolin 2
|
|
chr17_-_15164043
|
3.268
|
NM_153322
|
PMP22
|
peripheral myelin protein 22
|
|
chr15_-_23932410
|
3.187
|
NM_002487
|
NDN
|
necdin homolog (mouse)
|
|
chr7_+_116139619
|
3.134
|
NM_001206747
NM_001233
NM_198212
|
CAV2
|
caveolin 2
|
|
chr11_+_19734821
|
3.031
|
NM_001244963
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr5_-_111312522
|
2.990
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr12_-_6665220
|
2.968
|
NM_001039670
NM_001193457
NM_080730
|
IFFO1
|
intermediate filament family orphan 1
|
|
chr8_+_1772095
|
2.926
|
NM_014629
|
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chr1_+_151032150
|
2.894
|
NM_006818
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr1_-_225840660
|
2.877
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr5_-_111093792
|
2.848
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr2_+_9346876
|
2.777
|
NM_001135191
NM_003887
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr5_-_111093030
|
2.757
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr11_-_117166247
|
2.756
|
NM_001207048
NM_001207049
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr11_+_19372270
|
2.684
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr2_-_238322840
|
2.675
|
NM_004369
NM_057164
NM_057165
NM_057166
NM_057167
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr17_-_48278987
|
2.641
|
NM_000088
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr1_+_182992329
|
2.622
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chrX_-_51239426
|
2.591
|
NM_018159
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr5_-_111092918
|
2.569
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr3_-_149421059
|
2.567
|
NM_001168278
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr3_-_149375584
|
2.564
|
NM_001168280
NM_015472
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr19_+_48216600
|
2.563
|
NM_014601
|
EHD2
|
EH-domain containing 2
|
|
chr11_-_117186918
|
2.531
|
NM_012104
NM_138971
NM_138972
NM_138973
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr9_-_130639939
|
2.507
|
NM_000476
|
AK1
|
adenylate kinase 1
|
|
chr1_+_53068042
|
2.497
|
NM_015696
|
GPX6
GPX7
|
glutathione peroxidase 6 (olfactory)
glutathione peroxidase 7
|
|
chr5_-_111093251
|
2.493
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111091947
|
2.445
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr6_-_52860076
|
2.371
|
NM_001512
|
GSTA4
|
glutathione S-transferase alpha 4
|
|
chr11_+_75273100
|
2.366
|
NM_001207014
NM_001235
|
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1)
|
|
chr17_-_40575117
|
2.349
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr2_-_190927321
|
2.345
|
NM_005259
|
MSTN
|
myostatin
|
|
chr1_-_22263676
|
2.320
|
NM_005529
|
HSPG2
|
heparan sulfate proteoglycan 2
|
|
chr1_-_17308072
|
2.267
|
NM_001135247
NM_017459
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr10_-_735522
|
2.243
|
NM_014974
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila)
|
|
chr8_-_23261675
|
2.183
|
NM_002318
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr4_-_52904424
|
2.096
|
NM_000232
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein)
|
|
chr2_+_173292306
|
2.050
|
NM_000210
NM_001079818
|
ITGA6
|
integrin, alpha 6
|
|
chr1_-_17307107
|
1.994
|
NM_001135248
NM_002403
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr14_-_65438794
|
1.988
|
NM_198686
|
RAB15
|
RAB15, member RAS onocogene family
|
|
chr3_-_8811263
|
1.930
|
NM_000916
|
OXTR
|
oxytocin receptor
|
|
chrY_+_15016018
|
1.899
|
NM_001122665
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr9_-_14398981
|
1.823
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chrX_+_9754465
|
1.790
|
NM_001649
|
SHROOM2
|
shroom family member 2
|
|
chr16_+_69220940
|
1.760
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr11_+_844445
|
1.744
|
NM_001025237
|
TSPAN4
|
tetraspanin 4
|
|
chr6_-_52441815
|
1.707
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr12_-_6451156
|
1.698
|
NM_001065
|
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A
|
|
chr16_-_10674443
|
1.667
|
NM_001424
|
EMP2
|
epithelial membrane protein 2
|
|
chr5_-_139726173
|
1.649
|
NM_001945
|
HBEGF
|
heparin-binding EGF-like growth factor
|
|
chr10_-_33247178
|
1.630
|
NM_002211
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr3_+_158787040
|
1.628
|
NM_001197113
NM_001197114
NM_001042705
NM_001042706
NM_001197100
|
IQCJ-SCHIP1
IQCJ
|
IQCJ-SCHIP1 readthrough
IQ motif containing J
|
|
chr10_-_33246786
|
1.608
|
NM_133376
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr11_+_844062
|
1.601
|
NM_001025234
|
TSPAN4
|
tetraspanin 4
|
|
chr10_+_124145584
|
1.598
|
NM_001195608
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr1_-_236228476
|
1.594
|
NM_002508
|
NID1
|
nidogen 1
|
|
chr10_+_124134093
|
1.594
|
NM_001001974
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr6_+_39760148
|
1.586
|
NM_001201427
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr10_+_104503724
|
1.571
|
NM_001083913
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr9_-_14314036
|
1.564
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr11_-_86666211
|
1.549
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr7_+_73442426
|
1.544
|
NM_000501
NM_001081752
NM_001081753
NM_001081754
NM_001081755
|
ELN
|
elastin
|
|
chrY_+_15016698
|
1.518
|
NM_004660
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr8_+_26435339
|
1.508
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr17_+_26662547
|
1.502
|
NM_021137
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial)
|
|
chr17_-_55980749
|
1.502
|
NM_017949
|
CUEDC1
|
CUE domain containing 1
|
|
chr5_-_149535420
|
1.496
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr14_-_53417808
|
1.488
|
NM_001134999
NM_001135000
NM_006832
|
FERMT2
|
fermitin family member 2
|
|
chr11_+_842821
|
1.482
|
NM_001025238
NM_001025239
NM_003271
NM_001025235
NM_001025236
|
TSPAN4
|
tetraspanin 4
|
|
chr1_+_157963062
|
1.465
|
NM_018240
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr14_+_55034329
|
1.458
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr9_+_35161950
|
1.449
|
NM_006377
|
UNC13B
|
unc-13 homolog B (C. elegans)
|
|
chr12_-_51477299
|
1.428
|
NM_030809
|
CSRNP2
|
cysteine-serine-rich nuclear protein 2
|
|
chr6_-_134498973
|
1.421
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr7_+_12726413
|
1.404
|
NM_001037164
NM_212460
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr9_-_21335239
|
1.395
|
NM_018847
|
KLHL9
|
kelch-like 9 (Drosophila)
|
|
chr7_+_12726910
|
1.384
|
NM_001195396
NM_005738
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr6_-_134496006
|
1.381
|
NM_005627
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr6_+_143929316
|
1.374
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr1_-_36789754
|
1.369
|
NM_018166
|
FAM176B
|
family with sequence similarity 176, member B
|
|
chr6_+_143999058
|
1.366
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chrX_-_92928566
|
1.359
|
NM_004538
|
NAP1L3
|
nucleosome assembly protein 1-like 3
|
|
chr22_-_30642683
|
1.358
|
NM_002309
|
LIF
|
leukemia inhibitory factor (cholinergic differentiation factor)
|
|
chr6_+_39760772
|
1.358
|
NM_015345
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr6_-_134639195
|
1.355
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr7_+_128379345
|
1.346
|
NM_001130674
NM_001199671
NM_001199672
NM_001199673
NM_001199674
NM_001219
|
CALU
|
calumenin
|
|
chr10_+_124151818
|
1.343
|
NM_021622
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr7_+_106809405
|
1.309
|
NM_001244262
NM_012257
|
HBP1
|
HMG-box transcription factor 1
|
|
chr19_-_10121041
|
1.298
|
NM_015719
|
COL5A3
|
collagen, type V, alpha 3
|
|
chr12_-_65153189
|
1.290
|
NM_002076
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr1_-_227505796
|
1.284
|
NM_003607
NM_014826
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like)
|
|
chr8_+_26371461
|
1.283
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr8_-_93029864
|
1.235
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr10_-_30024594
|
1.235
|
NM_003174
|
SVIL
|
supervillin
|
|
chr12_+_3186520
|
1.234
|
NM_001168320
NM_006675
|
TSPAN9
|
tetraspanin 9
|
|
chr10_-_33224485
|
1.232
|
NM_033668
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr2_-_227663454
|
1.214
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr18_+_56530060
|
1.197
|
NM_018181
|
ZNF532
|
zinc finger protein 532
|
|
chr4_-_54930771
|
1.195
|
NM_012110
|
CHIC2
|
cysteine-rich hydrophobic domain 2
|
|
chr2_+_109237679
|
1.193
|
NM_001193484
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr22_-_36236421
|
1.190
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr6_-_134497069
|
1.186
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr22_-_36424397
|
1.179
|
NM_001082578
NM_001082579
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr4_+_124320664
|
1.165
|
NM_005841
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr6_-_110501164
|
1.163
|
NM_001024934
NM_001024935
NM_001024936
NM_003931
|
WASF1
|
WAS protein family, member 1
|
|
chr2_+_102314162
|
1.159
|
NM_001242560
NM_004834
NM_145686
NM_001242559
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr1_-_155881163
|
1.150
|
NM_006912
|
RIT1
|
Ras-like without CAAX 1
|
|
chr2_+_109271267
|
1.150
|
NM_001193485
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chrX_-_64254592
|
1.134
|
NM_001178032
NM_001243804
|
ZC4H2
|
zinc finger, C4H2 domain containing
|
|
chr17_-_7298160
|
1.128
|
NM_001201576
NM_020360
|
PLSCR3
|
phospholipid scramblase 3
|
|
chr10_+_75757847
|
1.125
|
NM_003373
NM_014000
|
VCL
|
vinculin
|
|
chr9_+_101705710
|
1.111
|
NM_001855
|
COL15A1
|
collagen, type XV, alpha 1
|
|
chrX_+_73641084
|
1.107
|
NM_006517
|
SLC16A2
|
solute carrier family 16, member 2 (monocarboxylic acid transporter 8)
|
|
chr2_+_109223600
|
1.105
|
NM_001193482
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr22_-_39636906
|
1.096
|
NM_033016
|
PDGFB
|
platelet-derived growth factor beta polypeptide
|
|
chr11_+_125034558
|
1.091
|
NM_022062
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr2_+_65283485
|
1.087
|
NM_015147
|
CEP68
|
centrosomal protein 68kDa
|
|
chrX_-_106960268
|
1.084
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr1_-_120612301
|
1.022
|
NM_001200001
NM_024408
|
NOTCH2
|
notch 2
|
|
chrX_+_49687212
|
1.015
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr9_+_112542496
|
0.981
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr4_+_124317884
|
0.980
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr2_+_109204745
|
0.976
|
NM_004987
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr1_+_203595897
|
0.972
|
NM_001001396
NM_001684
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr2_+_109150807
|
0.952
|
NM_001193483
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr1_+_218518675
|
0.952
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr2_+_109204904
|
0.948
|
NM_001193488
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr5_-_83016850
|
0.932
|
NM_001884
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chr9_+_112887780
|
0.930
|
NM_001136562
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr15_+_63334753
|
0.929
|
NM_000366
NM_001018004
NM_001018005
NM_001018006
NM_001018007
NM_001018020
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chrX_-_107019001
|
0.906
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr6_-_159065739
|
0.875
|
NM_006519
|
DYNLT1
|
dynein, light chain, Tctex-type 1
|
|
chr7_-_129592794
|
0.873
|
NM_003344
NM_182697
|
UBE2H
|
ubiquitin-conjugating enzyme E2H
|
|
chr10_+_115439425
|
0.871
|
NM_001227
NM_033338
NM_033339
|
CASP7
|
caspase 7, apoptosis-related cysteine peptidase
|
|
chr17_+_40169592
|
0.853
|
NM_001001349
|
NKIRAS2
|
NFKB inhibitor interacting Ras-like 2
|
|
chr9_+_112810877
|
0.844
|
NM_001004065
NM_001198656
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr10_-_32345337
|
0.844
|
NM_004521
|
KIF5B
|
kinesin family member 5B
|
|
chr3_+_43732358
|
0.837
|
NM_016006
|
ABHD5
|
abhydrolase domain containing 5
|
|
chr8_-_93111915
|
0.834
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr14_+_23067128
|
0.832
|
NM_022060
|
ABHD4
|
abhydrolase domain containing 4
|
|
chr8_-_93115453
|
0.830
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr20_-_39928708
|
0.829
|
NM_015035
|
ZHX3
|
zinc fingers and homeoboxes 3
|
|
chr7_+_17338182
|
0.829
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr17_+_40172086
|
0.818
|
NM_001144927
NM_001144928
NM_001144929
NM_017595
|
NKIRAS2
|
NFKB inhibitor interacting Ras-like 2
|
|
chr19_+_708766
|
0.816
|
NM_001040134
NM_002579
|
PALM
|
paralemmin
|
|
chr10_-_111683245
|
0.812
|
NM_001167604
NM_020383
|
XPNPEP1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble
|
|
chr6_-_56112315
|
0.806
|
NM_030820
|
COL21A1
|
collagen, type XXI, alpha 1
|
|
chr10_-_29923900
|
0.790
|
NM_021738
|
SVIL
|
supervillin
|
|
chr3_+_126707380
|
0.786
|
NM_032242
|
PLXNA1
|
plexin A1
|
|
chr12_+_122516555
|
0.783
|
NM_014938
|
MLXIP
|
MLX interacting protein
|
|
chr10_+_11059854
|
0.772
|
NM_006561
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr7_-_129592711
|
0.760
|
NM_001202498
|
UBE2H
|
ubiquitin-conjugating enzyme E2H
|
|
chr8_+_56015013
|
0.756
|
NM_052898
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4
|
|
chr4_-_114900802
|
0.749
|
NM_024590
|
ARSJ
|
arylsulfatase family, member J
|
|
chr11_-_66335958
|
0.740
|
NM_003793
|
CTSF
|
cathepsin F
|
|
chr2_-_96931744
|
0.738
|
NM_001193304
NM_017849
|
TMEM127
|
transmembrane protein 127
|
|
chr10_-_75634219
|
0.723
|
NM_001204492
NM_001222
NM_172169
NM_172170
NM_172171
NM_172173
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma
|
|
chr1_-_244006471
|
0.721
|
NM_005465
NM_181690
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma)
|
|
chr8_-_93107881
|
0.720
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_+_38585703
|
0.716
|
NM_001146216
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr12_+_6493221
|
0.706
|
NM_002342
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3)
|
|
chr14_+_77228132
|
0.706
|
NM_014909
|
VASH1
|
vasohibin 1
|
|
chr1_+_169075869
|
0.703
|
NM_001677
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr22_-_39640956
|
0.703
|
NM_002608
|
PDGFB
|
platelet-derived growth factor beta polypeptide
|
|
chr15_+_86220267
|
0.702
|
NM_144767
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr7_+_872137
|
0.693
|
NM_001130965
|
SUN1
|
Sad1 and UNC84 domain containing 1
|
|
chr14_+_69865380
|
0.688
|
NM_001252148
NM_001252150
NM_018375
|
SLC39A9
|
solute carrier family 39 (zinc transporter), member 9
|
|
chr14_-_78083109
|
0.683
|
NM_004863
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2
|
|
chr1_+_99729847
|
0.673
|
NM_001166252
NM_014839
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|