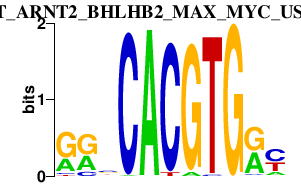
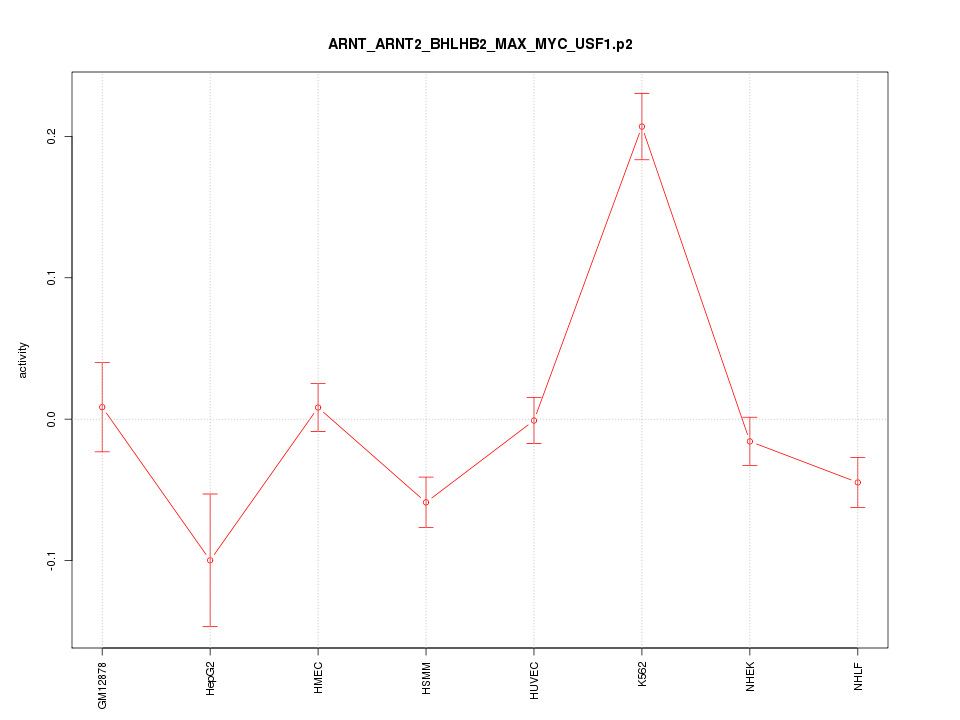
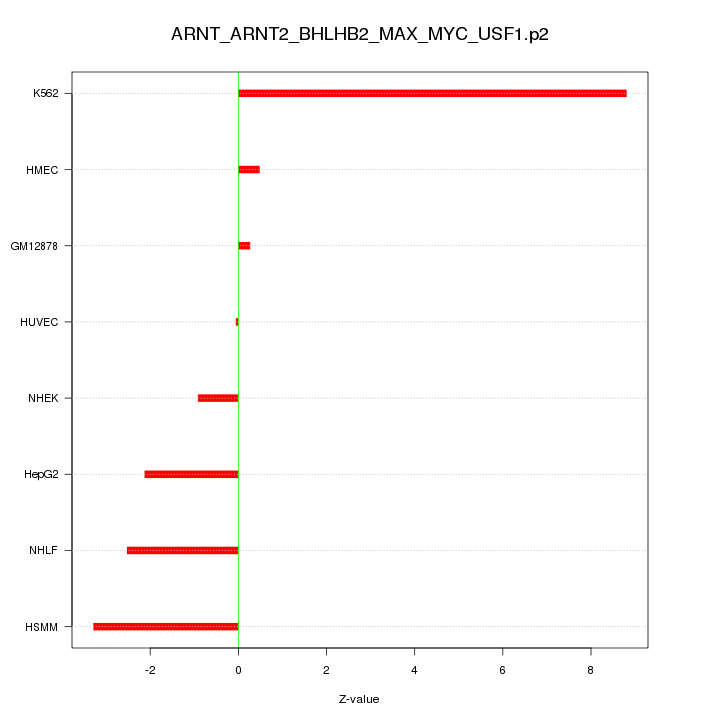
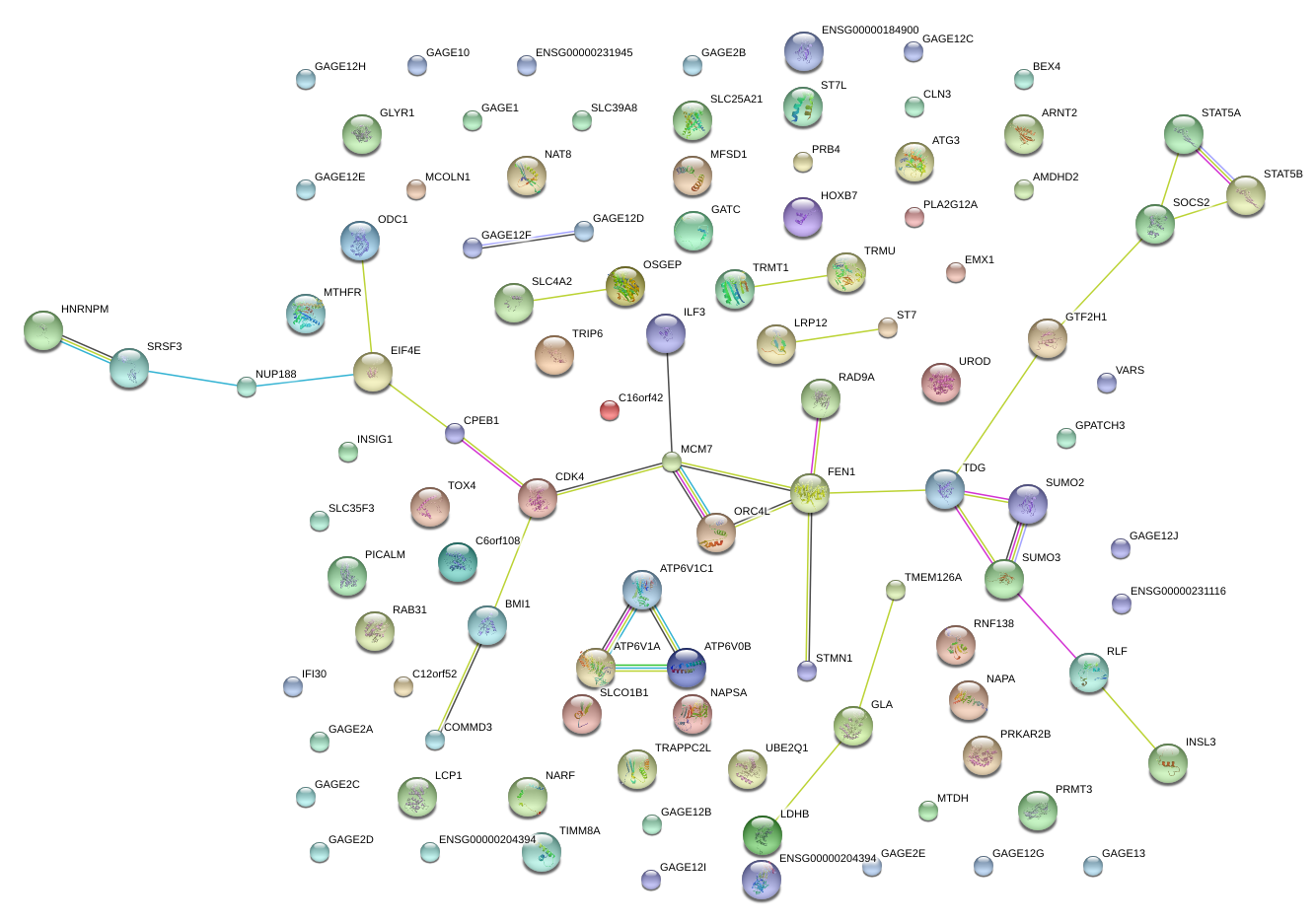
|
chr12_-_58146108
|
9.653
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr11_+_67159421
|
9.428
|
NM_004584
|
RAD9A
|
RAD9 homolog A (S. pombe)
|
|
chr12_-_58146025
|
9.264
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr12_-_58146110
|
8.482
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr12_-_58146118
|
8.171
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr16_+_2570322
|
7.591
|
NM_001145815
NM_015944
|
AMDHD2
|
amidohydrolase domain containing 2
|
|
chr1_-_26232890
|
6.920
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr4_-_103266577
|
6.908
|
NM_001135146
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8
|
|
chrX_+_102470003
|
6.885
|
NM_001080425
NM_001127688
|
BEX4
|
brain expressed, X-linked 4
|
|
chrX_-_100603956
|
6.486
|
NM_004085
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast)
|
|
chr14_-_20923266
|
5.826
|
NM_017807
|
OSGEP
|
O-sialoglycoprotein endopeptidase
|
|
chr6_-_43197178
|
5.799
|
NM_006443
NM_199184
|
C6orf108
|
chromosome 6 open reading frame 108
|
|
chr1_+_44440642
|
5.706
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr11_-_85780088
|
5.681
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr11_-_85780105
|
5.662
|
NM_001008660
NM_001206946
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr7_+_150759633
|
5.661
|
NM_001199693
NM_001199694
|
SLC4A2
|
solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1)
|
|
chr1_+_44440673
|
5.253
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr3_+_113465901
|
5.176
|
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr7_+_106685007
|
5.141
|
NM_002736
|
PRKAR2B
|
protein kinase, cAMP-dependent, regulatory, type II, beta
|
|
chr6_-_31763415
|
5.116
|
|
VARS
|
valyl-tRNA synthetase
|
|
chr1_+_44440623
|
5.113
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr18_+_9708269
|
5.013
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chrX_-_100662905
|
4.881
|
NM_000169
|
GLA
|
galactosidase, alpha
|
|
chrX_+_49235707
|
4.866
|
NM_001098411
|
GAGE2B
|
G antigen 2B
|
|
chr1_+_44440595
|
4.664
|
NM_001039457
NM_004047
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr9_+_131709991
|
4.662
|
|
NUP188
|
nucleoporin 188kDa
|
|
chr7_+_155089614
|
4.641
|
|
INSIG1
|
insulin induced gene 1
|
|
chrX_-_100662788
|
4.640
|
|
GLA
|
galactosidase, alpha
|
|
chr9_+_131709970
|
4.631
|
NM_015354
|
NUP188
|
nucleoporin 188kDa
|
|
chr18_+_9708322
|
4.618
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr7_-_99698993
|
4.604
|
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr18_+_9708299
|
4.594
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr6_-_33385814
|
4.523
|
|
|
|
|
chr7_-_99699030
|
4.501
|
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr19_-_48018277
|
4.484
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr3_+_158519888
|
4.445
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr11_+_18344113
|
4.369
|
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr11_+_61560108
|
4.312
|
NM_004111
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr4_-_103266396
|
4.291
|
NM_001135147
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8
|
|
chr11_-_85779821
|
4.258
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr6_-_31763473
|
4.217
|
|
VARS
|
valyl-tRNA synthetase
|
|
chr1_-_53164014
|
4.196
|
|
SELRC1
|
Sel1 repeat containing 1
|
|
chr7_+_155089483
|
4.184
|
NM_005542
NM_198336
NM_198337
|
INSIG1
|
insulin induced gene 1
|
|
chr16_-_4897265
|
4.176
|
NM_032569
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr7_-_99698916
|
4.087
|
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr19_+_10765106
|
4.069
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr17_+_40440481
|
4.006
|
|
STAT5A
|
signal transducer and activator of transcription 5A
|
|
chr1_-_26233367
|
3.957
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr1_+_45477888
|
3.898
|
|
UROD
|
uroporphyrinogen decarboxylase
|
|
chr19_+_18284631
|
3.823
|
|
IFI30
|
interferon, gamma-inducible protein 30
|
|
chr12_+_113623364
|
3.765
|
NM_032848
|
C12orf52
|
chromosome 12 open reading frame 52
|
|
chr16_+_88923490
|
3.690
|
NM_016209
|
TRAPPC2L
|
trafficking protein particle complex 2-like
|
|
chrX_+_49296768
|
3.681
|
NM_001127199
NM_001098409
NM_001098408
NM_001127345
NM_001098418
NM_001098410
|
GAGE12D
GAGE6
GAGE7
GAGE12G
GAGE12C
GAGE12B
GAGE12E
GAGE12H
|
G antigen 12D
G antigen 6
G antigen 7
G antigen 12G
G antigen 12C
G antigen 12B
G antigen 12E
G antigen 12H
|
|
chr18_+_9708187
|
3.660
|
NM_006868
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chrX_+_49188092
|
3.654
|
NM_012196
NM_001127200
NM_001098412
|
GAGE8
GAGE2C
GAGE2E
GAGE13
GAGE2A
|
G antigen 8
G antigen 2C
G antigen 2E
G antigen 13
G antigen 2A
|
|
chr19_+_7587436
|
3.634
|
NM_020533
|
MCOLN1
|
mucolipin 1
|
|
chr1_-_154531083
|
3.628
|
NM_017582
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr12_+_93964801
|
3.618
|
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr19_+_18284619
|
3.595
|
|
IFI30
|
interferon, gamma-inducible protein 30
|
|
chrX_+_49306324
|
3.588
|
NM_001127199
NM_001098409
NM_001098408
NM_001127345
NM_001098418
NM_001098410
|
GAGE12D
GAGE6
GAGE7
GAGE12G
GAGE12C
GAGE12B
GAGE12E
GAGE12H
|
G antigen 12D
G antigen 6
G antigen 7
G antigen 12G
G antigen 12C
G antigen 12B
G antigen 12E
G antigen 12H
|
|
chr19_+_18284578
|
3.572
|
NM_006332
|
IFI30
|
interferon, gamma-inducible protein 30
|
|
chr15_-_83316621
|
3.568
|
NM_030594
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr12_+_104359557
|
3.555
|
NM_003211
|
TDG
|
thymine-DNA glycosylase
|
|
chr11_+_18343794
|
3.548
|
NM_001142307
NM_005316
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr11_+_61560401
|
3.532
|
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr14_-_20922969
|
3.489
|
|
OSGEP
|
O-sialoglycoprotein endopeptidase
|
|
chr2_-_10588451
|
3.446
|
NM_002539
|
ODC1
|
ornithine decarboxylase 1
|
|
chr16_-_28503079
|
3.437
|
NM_000086
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3
|
|
chrX_+_49216616
|
3.408
|
NM_012196
NM_001472
NM_001474
NM_001475
NM_021123
NM_001477
NM_001127200
NM_001127212
|
GAGE8
GAGE1
GAGE2C
GAGE4
GAGE5
GAGE6
GAGE7
GAGE12I
GAGE2E
GAGE12B
GAGE2A
|
G antigen 8
G antigen 1
G antigen 2C
G antigen 4
G antigen 5
G antigen 6
G antigen 7
G antigen 12I
G antigen 2E
G antigen 12B
G antigen 2A
|
|
chr19_-_48018317
|
3.370
|
NM_003827
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chrX_+_49344540
|
3.356
|
NM_001127199
NM_001098408
NM_001098418
NM_001098410
|
GAGE12D
GAGE12C
GAGE12B
GAGE12E
GAGE12H
|
G antigen 12D
G antigen 12C
G antigen 12B
G antigen 12E
G antigen 12H
|
|
chr6_+_36562135
|
3.351
|
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr4_-_99850242
|
3.308
|
NM_001130678
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chr1_-_26232643
|
3.288
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr1_+_45477916
|
3.286
|
|
UROD
|
uroporphyrinogen decarboxylase
|
|
chr19_-_13227212
|
3.286
|
NM_001142554
NM_017722
NM_001136035
|
TRMT1
|
TRM1 tRNA methyltransferase 1 homolog (S. cerevisiae)
|
|
chr1_-_53163985
|
3.222
|
|
SELRC1
|
Sel1 repeat containing 1
|
|
chr3_+_113465860
|
3.098
|
NM_001690
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr1_-_27226928
|
3.069
|
|
GPATCH3
|
G patch domain containing 3
|
|
chrX_+_49207115
|
3.037
|
NM_012196
NM_001472
NM_001127200
NM_001098407
NM_001127212
|
GAGE8
GAGE2C
GAGE2E
GAGE2D
GAGE2A
|
G antigen 8
G antigen 2C
G antigen 2E
G antigen 2D
G antigen 2A
|
|
chr11_+_61560352
|
3.013
|
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr21_-_46237974
|
2.986
|
|
SUMO3
|
SMT3 suppressor of mif two 3 homolog 3 (S. cerevisiae)
|
|
chr19_+_10764987
|
2.968
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr14_-_20922841
|
2.962
|
|
OSGEP
|
O-sialoglycoprotein endopeptidase
|
|
chr1_+_45477801
|
2.950
|
NM_000374
|
UROD
|
uroporphyrinogen decarboxylase
|
|
chr19_-_48018163
|
2.946
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr12_-_21810779
|
2.897
|
NM_002300
|
LDHB
|
lactate dehydrogenase B
|
|
chr12_-_21810720
|
2.879
|
NM_001174097
|
LDHB
|
lactate dehydrogenase B
|
|
chr7_+_116593554
|
2.871
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr4_-_99850280
|
2.853
|
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chr21_-_46237978
|
2.845
|
|
SUMO3
|
SMT3 suppressor of mif two 3 homolog 3 (S. cerevisiae)
|
|
chr12_-_21810733
|
2.831
|
|
LDHB
|
lactate dehydrogenase B
|
|
chrX_+_49315880
|
2.815
|
NM_001127199
NM_001098409
NM_001098408
NM_001127345
NM_001098418
NM_001098410
|
GAGE12D
GAGE6
GAGE7
GAGE12G
GAGE12C
GAGE12B
GAGE12E
GAGE12H
|
G antigen 12D
G antigen 6
G antigen 7
G antigen 12G
G antigen 12C
G antigen 12B
G antigen 12E
G antigen 12H
|
|
chrX_+_49197555
|
2.814
|
NM_012196
NM_001472
NM_001127200
NM_001098407
NM_001127212
|
GAGE8
GAGE2C
GAGE2E
GAGE12J
GAGE2D
GAGE2A
|
G antigen 8
G antigen 2C
G antigen 2E
G antigen 12J
G antigen 2D
G antigen 2A
|
|
chr13_-_46742678
|
2.792
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr16_-_1401765
|
2.781
|
NM_001001410
|
C16orf42
|
chromosome 16 open reading frame 42
|
|
chr11_+_20409075
|
2.779
|
NM_001145166
NM_001145167
NM_005788
|
PRMT3
|
protein arginine methyltransferase 3
|
|
chr17_+_80416047
|
2.778
|
NM_001038618
|
NARF
|
nuclear prelamin A recognition factor
|
|
chr1_+_40627066
|
2.759
|
|
RLF
|
rearranged L-myc fusion
|
|
chr12_-_21810754
|
2.754
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr8_+_98656428
|
2.748
|
|
MTDH
|
metadherin
|
|
chr19_+_8509869
|
2.745
|
|
HNRNPM
|
heterogeneous nuclear ribonucleoprotein M
|
|
chr12_-_21810769
|
2.745
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr17_-_46688362
|
2.743
|
NM_004502
|
HOXB7
|
homeobox B7
|
|
chr3_-_112280764
|
2.733
|
|
ATG3
|
ATG3 autophagy related 3 homolog (S. cerevisiae)
|
|
chr10_+_22605300
|
2.663
|
NM_001204062
NM_012071
|
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough
COMM domain containing 3
|
|
chr2_-_148778262
|
2.640
|
NM_001190882
NM_001190879
NM_002552
NM_001190881
NM_181741
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr1_-_11866079
|
2.637
|
NM_005957
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr7_+_100464949
|
2.627
|
NM_003302
|
TRIP6
|
thyroid hormone receptor interactor 6
|
|
chr8_+_104033301
|
2.614
|
|
ATP6V1C1
|
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1
|
|
chr19_+_8509802
|
2.594
|
NM_005968
NM_031203
|
HNRNPM
|
heterogeneous nuclear ribonucleoprotein M
|
|
chr4_-_110650965
|
2.583
|
NM_030821
|
PLA2G12A
|
phospholipase A2, group XIIA
|
|
chr1_-_111991790
|
2.572
|
NM_024102
|
WDR77
|
WD repeat domain 77
|
|
chr17_-_41132019
|
2.565
|
NM_025267
|
AARSD1
|
alanyl-tRNA synthetase domain containing 1
|
|
chr19_+_10812111
|
2.565
|
NM_031209
|
QTRT1
|
queuine tRNA-ribosyltransferase 1
|
|
chr1_-_202896315
|
2.560
|
|
KLHL12
|
kelch-like 12 (Drosophila)
|
|
chr1_+_40627031
|
2.541
|
NM_012421
|
RLF
|
rearranged L-myc fusion
|
|
chr1_-_53164023
|
2.518
|
NM_023077
|
SELRC1
|
Sel1 repeat containing 1
|
|
chr17_+_46970128
|
2.501
|
NM_001002027
NM_005175
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9)
|
|
chr1_+_26798901
|
2.500
|
NM_005517
|
HMGN2
|
high mobility group nucleosomal binding domain 2
|
|
chr2_-_220042731
|
2.471
|
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1
|
|
chr18_+_32556891
|
2.448
|
NM_001143826
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr11_-_118972529
|
2.427
|
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase)
|
|
chr17_-_46688279
|
2.421
|
|
HOXB7
|
homeobox B7
|
|
chr11_-_118972784
|
2.419
|
NM_001382
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase)
|
|
chr1_-_11120081
|
2.399
|
NM_003132
|
SRM
|
spermidine synthase
|
|
chr17_-_47841517
|
2.385
|
NM_030802
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr6_-_31763551
|
2.382
|
NM_006295
|
VARS
|
valyl-tRNA synthetase
|
|
chr11_+_20409212
|
2.341
|
|
PRMT3
|
protein arginine methyltransferase 3
|
|
chr1_-_111991859
|
2.330
|
|
WDR77
|
WD repeat domain 77
|
|
chr16_+_2867163
|
2.329
|
NM_006799
NM_144956
NM_144957
|
PRSS21
|
protease, serine, 21 (testisin)
|
|
chr12_+_132413776
|
2.318
|
NM_001002019
NM_025215
|
PUS1
|
pseudouridylate synthase 1
|
|
chr1_-_159895279
|
2.292
|
NM_003564
|
TAGLN2
|
transgelin 2
|
|
chrX_-_49460568
|
2.283
|
NM_003785
|
PAGE1
|
P antigen family, member 1 (prostate associated)
|
|
chr1_+_182758871
|
2.272
|
|
NPL
|
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase)
|
|
chrX_+_49354117
|
2.267
|
NM_001127212
|
GAGE1
GAGE2C
GAGE2A
|
G antigen 1
G antigen 2C
G antigen 2A
|
|
chr2_+_27440342
|
2.254
|
|
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase
|
|
chr1_-_27226937
|
2.243
|
NM_022078
|
GPATCH3
|
G patch domain containing 3
|
|
chr9_-_116172658
|
2.232
|
|
POLE3
|
polymerase (DNA directed), epsilon 3 (p17 subunit)
|
|
chr5_+_89770680
|
2.204
|
NM_006467
|
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)
|
|
chr1_+_10092990
|
2.204
|
NM_001105562
NM_006048
|
UBE4B
|
ubiquitination factor E4B
|
|
chr13_-_36788665
|
2.203
|
NM_017826
|
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2
|
|
chr17_+_49337882
|
2.195
|
|
UTP18
|
UTP18 small subunit (SSU) processome component homolog (yeast)
|
|
chr3_+_113465910
|
2.190
|
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr16_-_89043215
|
2.182
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr16_+_2867227
|
2.174
|
|
PRSS21
|
protease, serine, 21 (testisin)
|
|
chr3_-_170626425
|
2.172
|
NM_020390
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr1_-_11120047
|
2.170
|
|
SRM
|
spermidine synthase
|
|
chr7_-_99699426
|
2.170
|
NM_005916
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr5_+_70883132
|
2.170
|
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr8_+_98656329
|
2.166
|
NM_178812
|
MTDH
|
metadherin
|
|
chr3_+_158519911
|
2.155
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr1_-_229761793
|
2.144
|
NM_001025247
NM_014409
|
TAF5L
|
TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa
|
|
chrX_+_49226137
|
2.137
|
NM_001472
NM_001098407
|
GAGE2C
GAGE2E
GAGE2D
GAGE2A
|
G antigen 2C
G antigen 2E
G antigen 2D
G antigen 2A
|
|
chr19_+_797444
|
2.134
|
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chr10_-_12085022
|
2.123
|
NM_080599
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast)
|
|
chr5_-_131132642
|
2.117
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr16_+_68119211
|
2.109
|
NM_004555
NM_173163
NM_173165
|
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3
|
|
chrX_+_100663205
|
2.106
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr6_-_28891409
|
2.094
|
|
TRIM27
|
tripartite motif containing 27
|
|
chr6_+_7590405
|
2.089
|
NM_152551
|
SNRNP48
|
small nuclear ribonucleoprotein 48kDa (U11/U12)
|
|
chr8_+_98656494
|
2.084
|
|
MTDH
|
metadherin
|
|
chr11_+_66624501
|
2.071
|
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr1_+_10093149
|
2.068
|
|
UBE4B
|
ubiquitination factor E4B
|
|
chr3_+_184032414
|
2.066
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr12_-_122750954
|
2.066
|
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae)
|
|
chr4_-_103266286
|
2.061
|
NM_022154
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8
|
|
chr17_+_49337909
|
2.026
|
|
UTP18
|
UTP18 small subunit (SSU) processome component homolog (yeast)
|
|
chr1_+_154193757
|
2.014
|
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chrX_+_118108575
|
2.011
|
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr1_-_202896368
|
2.011
|
NM_021633
|
KLHL12
|
kelch-like 12 (Drosophila)
|
|
chr10_-_43903220
|
1.998
|
NM_001098206
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F
|
|
chrX_+_16804531
|
1.992
|
NM_001168683
NM_018360
|
TXLNG
|
taxilin gamma
|
|
chr3_+_50654589
|
1.991
|
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr17_+_49337892
|
1.986
|
NM_016001
|
UTP18
|
UTP18 small subunit (SSU) processome component homolog (yeast)
|
|
chr2_+_68384986
|
1.983
|
NM_020143
|
PNO1
|
partner of NOB1 homolog (S. cerevisiae)
|
|
chr16_-_1525020
|
1.980
|
|
CLCN7
|
chloride channel 7
|
|
chrX_+_64887510
|
1.970
|
NM_002444
|
MSN
|
moesin
|
|
chrX_+_131157231
|
1.953
|
NM_001042453
NM_016542
|
MST4
|
serine/threonine protein kinase MST4
|
|
chrX_+_49178508
|
1.946
|
NM_001098406
|
GAGE1
GAGE4
GAGE5
GAGE6
GAGE12J
|
G antigen 1
G antigen 4
G antigen 5
G antigen 6
G antigen 12J
|
|
chr7_+_102074002
|
1.943
|
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr12_-_122751044
|
1.940
|
NM_022916
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae)
|
|
chr19_+_1275510
|
1.926
|
NM_017914
|
C19orf24
|
chromosome 19 open reading frame 24
|
|
chr1_+_40627055
|
1.919
|
|
RLF
|
rearranged L-myc fusion
|
|
chr1_-_159895258
|
1.917
|
|
TAGLN2
|
transgelin 2
|
|
chrX_+_49325433
|
1.916
|
NM_001127199
NM_001476
NM_001477
NM_001098409
NM_001098408
NM_001098418
|
GAGE12D
GAGE1
GAGE4
GAGE5
GAGE6
GAGE7
GAGE12I
GAGE12G
GAGE12C
GAGE12B
GAGE12E
|
G antigen 12D
G antigen 1
G antigen 4
G antigen 5
G antigen 6
G antigen 7
G antigen 12I
G antigen 12G
G antigen 12C
G antigen 12B
G antigen 12E
|
|
chr6_-_106773290
|
1.916
|
|
ATG5
|
ATG5 autophagy related 5 homolog (S. cerevisiae)
|
|
chr3_+_184032319
|
1.911
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr6_-_106773607
|
1.911
|
NM_004849
|
ATG5
|
ATG5 autophagy related 5 homolog (S. cerevisiae)
|
|
chr1_+_26798961
|
1.910
|
|
HMGN2
|
high mobility group nucleosomal binding domain 2
|
|
chr16_-_1525006
|
1.909
|
|
CLCN7
|
chloride channel 7
|
|
chr17_+_35306174
|
1.907
|
NM_012138
|
AATF
|
apoptosis antagonizing transcription factor
|
|
chr5_+_133706869
|
1.902
|
NM_003337
|
UBE2B
|
ubiquitin-conjugating enzyme E2B
|
|
chr9_-_103115094
|
1.897
|
NM_017746
|
TEX10
|
testis expressed 10
|
|
chr17_-_63052816
|
1.892
|
|
|
|
|
chr17_-_63052884
|
1.882
|
NM_006572
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chrX_+_49334991
|
1.882
|
NM_001127199
NM_001476
NM_001477
NM_001098409
NM_001098408
NM_001098418
|
GAGE12D
GAGE1
GAGE4
GAGE5
GAGE6
GAGE7
GAGE12I
GAGE12G
GAGE12C
GAGE12B
GAGE12E
|
G antigen 12D
G antigen 1
G antigen 4
G antigen 5
G antigen 6
G antigen 7
G antigen 12I
G antigen 12G
G antigen 12C
G antigen 12B
G antigen 12E
|
|
chr10_+_75936220
|
1.876
|
NM_001123
NM_001202449
|
ADK
|
adenosine kinase
|
|
chr2_+_216176808
|
1.867
|
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr1_-_28559513
|
1.863
|
NM_014280
|
DNAJC8
|
DnaJ (Hsp40) homolog, subfamily C, member 8
|