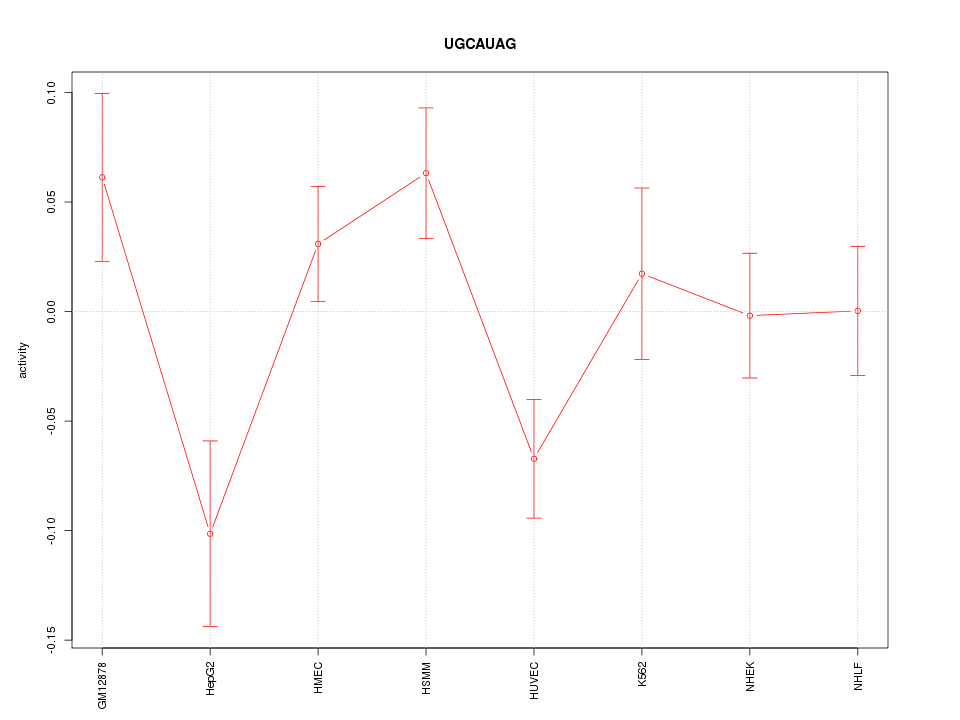
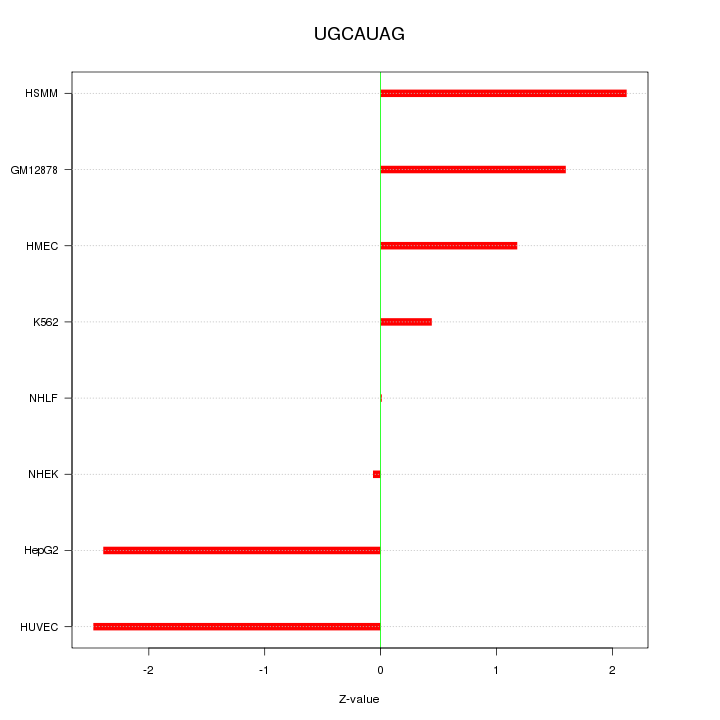
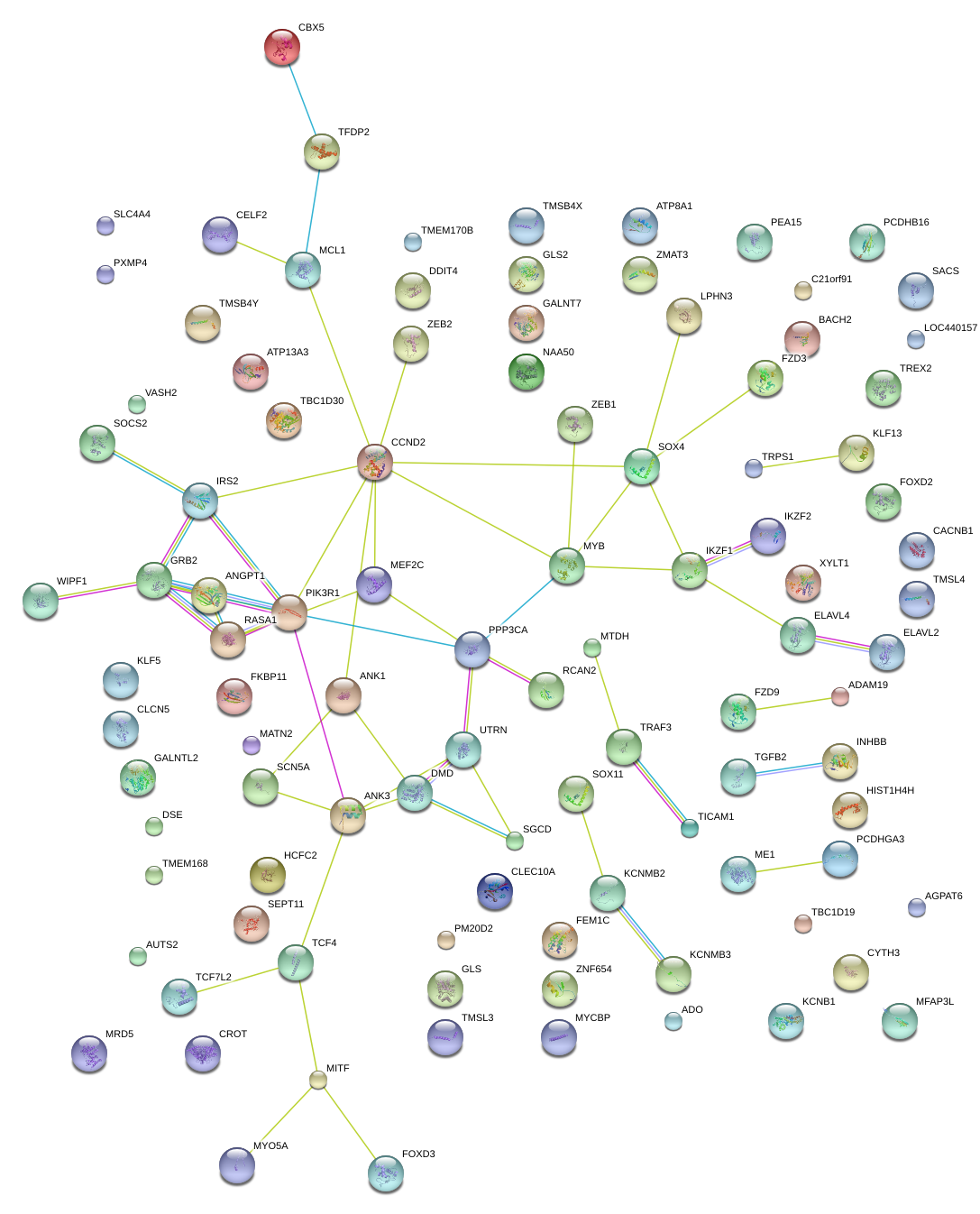
|
chr4_+_174089889
|
2.605
|
NM_017423
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7)
|
|
chr7_+_69063768
|
1.974
|
NM_001127231
NM_001127232
NM_015570
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr6_+_116692098
|
1.946
|
NM_013352
|
DSE
|
dermatan sulfate epimerase
|
|
chr12_+_4382882
|
1.934
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr6_+_116601282
|
1.920
|
NM_001080976
|
DSE
|
dermatan sulfate epimerase
|
|
chr1_+_213123853
|
1.792
|
NM_001136474
NM_001136475
NM_024749
|
VASH2
|
vasohibin 2
|
|
chrX_+_12993224
|
1.730
|
NM_021109
|
TMSB4X
|
thymosin beta 4, X-linked
|
|
chr5_-_157002739
|
1.688
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr5_-_88199868
|
1.656
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr13_-_110438896
|
1.589
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr5_-_88179278
|
1.391
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr10_+_11059854
|
1.360
|
NM_006561
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr10_+_11047253
|
1.344
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr16_-_17564737
|
1.314
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr10_+_11206928
|
1.278
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr20_+_9494995
|
1.233
|
NM_001199897
NM_012261
|
LAMP5
|
lysosomal-associated membrane protein family, member 5
|
|
chr2_-_175547471
|
1.214
|
NM_001077269
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr2_-_175499275
|
1.148
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr1_-_39338976
|
1.137
|
NM_012333
|
MYCBP
|
c-myc binding protein
|
|
chr2_+_5832778
|
1.100
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr21_-_19191643
|
1.075
|
NM_001100420
NM_001100421
NM_017447
|
C21orf91
|
chromosome 21 open reading frame 91
|
|
chr3_+_88188261
|
1.062
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr5_-_88119604
|
1.050
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr2_-_145277879
|
1.041
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr8_-_108510094
|
1.021
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr1_+_160175114
|
1.020
|
NM_003768
|
PEA15
|
phosphoprotein enriched in astrocytes 15
|
|
chr6_-_84140854
|
1.018
|
NM_002395
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr15_-_52821004
|
1.017
|
NM_000259
NM_001142495
|
MYO5A
|
myosin VA (heavy chain 12, myoxin)
|
|
chr3_+_69812620
|
0.977
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr12_+_93963597
|
0.972
|
NM_003877
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr8_-_41754244
|
0.932
|
NM_001142446
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr20_-_32307889
|
0.925
|
NM_007238
NM_183397
|
PXMP4
|
peroxisomal membrane protein 4, 24kDa
|
|
chr4_+_77870864
|
0.873
|
NM_018243
|
SEPT11
|
septin 11
|
|
chr7_+_50344255
|
0.871
|
NM_001220765
NM_001220766
NM_001220767
NM_001220768
NM_001220769
NM_001220770
NM_001220771
NM_001220772
NM_001220773
NM_001220774
NM_001220775
NM_001220776
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr2_+_191745546
|
0.863
|
NM_014905
|
GLS
|
glutaminase
|
|
chr10_+_74033676
|
0.819
|
NM_019058
|
DDIT4
|
DNA-damage-inducible transcript 4
|
|
chr3_+_69812961
|
0.786
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr8_-_116681103
|
0.771
|
NM_014112
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr8_+_28351694
|
0.767
|
NM_017412
NM_145866
|
FZD3
|
frizzled family receptor 3
|
|
chrX_+_49832214
|
0.765
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr6_+_135502445
|
0.761
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr15_+_31619043
|
0.751
|
NM_015995
|
KLF13
|
Kruppel-like factor 13
|
|
chr8_-_41655139
|
0.750
|
NM_000037
NM_020475
NM_020476
NM_020477
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr3_-_141719188
|
0.738
|
NM_001178140
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr4_+_26585545
|
0.730
|
NM_018317
|
TBC1D19
|
TBC1 domain family, member 19
|
|
chr5_-_72744351
|
0.711
|
NM_004472
|
FOXD1
|
forkhead box D1
|
|
chr8_-_41522724
|
0.696
|
NM_001142445
NM_020478
NM_020480
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr12_+_65218351
|
0.686
|
NM_015279
|
TBC1D30
|
TBC1 domain family, member 30
|
|
chr10_-_62493282
|
0.641
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr1_+_218518675
|
0.639
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr9_-_23821477
|
0.632
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr20_-_48099178
|
0.632
|
NM_004975
|
KCNB1
|
potassium voltage-gated channel, Shab-related subfamily, member 1
|
|
chr3_-_38691118
|
0.629
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr3_-_141868208
|
0.624
|
NM_001178138
NM_001178139
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chrX_-_33229428
|
0.612
|
NM_004006
|
DMD
|
dystrophin
|
|
chr13_+_73633097
|
0.606
|
NM_001730
|
KLF5
|
Kruppel-like factor 5 (intestinal)
|
|
chr4_-_170924911
|
0.605
|
NM_001009554
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr5_+_155753755
|
0.601
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr7_+_86974928
|
0.595
|
NM_001143935
NM_001243745
NM_021151
|
CROT
|
carnitine O-octanoyltransferase
|
|
chr5_+_140797243
|
0.589
|
NM_018927
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr4_+_72052354
|
0.582
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr9_-_23821842
|
0.574
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr4_-_170947428
|
0.573
|
NM_021647
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr2_-_214015053
|
0.572
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr4_-_42658883
|
0.571
|
NM_001105529
NM_006095
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr4_-_102268626
|
0.545
|
NM_000944
NM_001130691
NM_001130692
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr3_+_69788562
|
0.539
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr8_+_98881277
|
0.527
|
NM_002380
NM_030583
|
MATN2
|
matrilin 2
|
|
chr14_+_103243813
|
0.526
|
NM_001199427
NM_003300
NM_145725
NM_145726
|
TRAF3
|
TNF receptor-associated factor 3
|
|
chr12_+_104458235
|
0.501
|
NM_013320
|
HCFC2
|
host cell factor C2
|
|
chr3_-_141747434
|
0.498
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr2_-_214016332
|
0.492
|
NM_001079526
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr3_-_178789597
|
0.489
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr6_+_11538486
|
0.482
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr10_+_64564480
|
0.478
|
NM_032804
|
ADO
|
2-aminoethanethiol (cysteamine) dioxygenase
|
|
chr6_-_91006460
|
0.471
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr6_-_46293628
|
0.467
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr8_+_98656329
|
0.466
|
NM_178812
|
MTDH
|
metadherin
|
|
chr18_-_53255734
|
0.464
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr5_-_114880536
|
0.462
|
NM_020177
|
FEM1C
|
fem-1 homolog c (C. elegans)
|
|
chr10_-_62149487
|
0.460
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr9_-_23826062
|
0.458
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr3_-_113465101
|
0.457
|
NM_025146
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr5_+_86564727
|
0.455
|
NM_022650
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr3_-_194188967
|
0.446
|
NM_024524
|
ATP13A3
|
ATPase type 13A3
|
|
chr5_+_67511578
|
0.445
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr13_-_24007822
|
0.437
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr5_+_86564044
|
0.436
|
NM_002890
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr10_-_61900659
|
0.433
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr7_-_112430408
|
0.433
|
NM_022484
|
TMEM168
|
transmembrane protein 168
|
|
chr3_+_69985750
|
0.430
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr17_-_73401788
|
0.429
|
NM_002086
NM_203506
|
GRB2
|
growth factor receptor-bound protein 2
|
|
chr3_+_178276487
|
0.428
|
NM_005832
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chr5_+_67588395
|
0.428
|
NM_001242466
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr10_-_62332666
|
0.426
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr1_-_150552127
|
0.424
|
NM_001197320
NM_021960
NM_182763
|
MCL1
|
myeloid cell leukemia sequence 1 (BCL2-related)
|
|
chr3_+_178254223
|
0.424
|
NM_181361
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chr7_-_6312137
|
0.423
|
NM_004227
|
CYTH3
|
cytohesin 3
|
|
chr2_+_121103670
|
0.423
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr12_-_54673901
|
0.421
|
NM_012117
|
CBX5
|
chromobox homolog 5
|
|
chr17_-_4269814
|
0.415
|
NM_003342
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1
|
|
chr2_-_171923482
|
0.413
|
NM_001136555
|
TLK1
|
tousled-like kinase 1
|
|
chr12_-_85306579
|
0.410
|
NM_001146335
NM_018057
NM_182767
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr5_-_179780314
|
0.407
|
NM_005110
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2
|
|
chr7_-_559479
|
0.406
|
NM_002607
NM_033023
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr4_-_5894695
|
0.402
|
NM_001014809
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr22_+_40440664
|
0.401
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr3_+_69915374
|
0.399
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr5_+_140749830
|
0.397
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr12_-_54653317
|
0.397
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chrX_+_44732402
|
0.396
|
NM_021140
|
KDM6A
|
lysine (K)-specific demethylase 6A
|
|
chr5_+_67586466
|
0.390
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr8_-_70745624
|
0.390
|
NM_001146008
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr12_+_111843743
|
0.390
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr3_-_160283361
|
0.390
|
NM_002268
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3)
|
|
chr2_-_1748285
|
0.389
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr1_+_100435539
|
0.386
|
NM_012243
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3
|
|
chrX_-_33146543
|
0.385
|
NM_004010
NM_004009
|
DMD
|
dystrophin
|
|
chr7_+_28452143
|
0.376
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr4_-_53522756
|
0.374
|
NM_001134223
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr8_-_95274540
|
0.366
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr5_+_140864626
|
0.365
|
NM_018928
NM_032406
|
PCDHGC4
|
protocadherin gamma subfamily C, 4
|
|
chr7_+_28725597
|
0.363
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr5_+_140810126
|
0.363
|
NM_003735
NM_032094
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr1_+_15736387
|
0.360
|
NM_024329
|
EFHD2
|
EF-hand domain family, member D2
|
|
chr14_+_66974124
|
0.358
|
NM_001024218
NM_020806
|
GPHN
|
gephyrin
|
|
chr5_+_140743755
|
0.357
|
NM_018918
NM_032054
|
PCDHGA5
|
protocadherin gamma subfamily A, 5
|
|
chr2_-_222436996
|
0.356
|
NM_004438
|
EPHA4
|
EPH receptor A4
|
|
chr16_+_82660390
|
0.350
|
NM_001220488
NM_001220489
NM_001220490
NM_001220491
NM_001220492
NM_001257
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr8_-_70745403
|
0.349
|
NM_001146009
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr1_+_110091185
|
0.349
|
NM_006496
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3
|
|
chr1_-_38325224
|
0.348
|
NM_005955
|
MTF1
|
metal-regulatory transcription factor 1
|
|
chr1_+_204913353
|
0.348
|
NM_001160331
|
NFASC
|
neurofascin
|
|
chr12_-_54653369
|
0.344
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr11_-_79151694
|
0.342
|
NM_001098816
|
ODZ4
|
odz, odd Oz/ten-m homolog 4 (Drosophila)
|
|
chr12_+_78225068
|
0.342
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr5_+_112312392
|
0.342
|
NM_001242377
NM_152624
|
DCP2
|
DCP2 decapping enzyme homolog (S. cerevisiae)
|
|
chr6_-_89673347
|
0.341
|
NM_003800
|
RNGTT
|
RNA guanylyltransferase and 5'-phosphatase
|
|
chr5_+_140868721
|
0.340
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr5_-_137071703
|
0.337
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr9_-_98279246
|
0.336
|
NM_001083602
NM_001083603
|
PTCH1
|
patched 1
|
|
chr18_-_23670533
|
0.336
|
NM_001007559
NM_005637
|
SS18
|
synovial sarcoma translocation, chromosome 18
|
|
chr5_+_140729723
|
0.336
|
NM_018922
NM_032095
|
PCDHGB1
|
protocadherin gamma subfamily B, 1
|
|
chr8_-_103876390
|
0.334
|
NM_015878
NM_148174
|
AZIN1
|
antizyme inhibitor 1
|
|
chr13_+_25875665
|
0.332
|
NM_001008564
NM_014089
|
NUPL1
|
nucleoporin like 1
|
|
chr4_+_140222668
|
0.330
|
NM_057175
|
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit
|
|
chr8_-_74884142
|
0.328
|
NM_001204861
NM_001204862
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C)
|
|
chr6_+_44238479
|
0.328
|
NM_001137560
|
TMEM151B
|
transmembrane protein 151B
|
|
chr10_-_21186530
|
0.324
|
NM_006393
|
NEBL
|
nebulette
|
|
chr6_-_91006580
|
0.322
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr5_+_140718353
|
0.316
|
NM_018915
NM_032009
|
PCDHGA2
|
protocadherin gamma subfamily A, 2
|
|
chr3_-_127541160
|
0.314
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr3_+_111717585
|
0.314
|
NM_001008272
NM_013259
|
TAGLN3
|
transgelin 3
|
|
chr10_-_119805673
|
0.312
|
NM_014904
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I)
|
|
chr9_+_129567260
|
0.309
|
NM_001135776
NM_014007
|
ZBTB43
|
zinc finger and BTB domain containing 43
|
|
chr9_-_95055966
|
0.306
|
NM_013417
|
IARS
|
isoleucyl-tRNA synthetase
|
|
chr7_+_28475233
|
0.304
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr5_+_140767402
|
0.299
|
NM_003736
NM_032098
|
PCDHGB4
|
protocadherin gamma subfamily B, 4
|
|
chr7_+_28338939
|
0.299
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr11_-_83393436
|
0.297
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chrX_+_49687212
|
0.295
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr11_+_125034558
|
0.291
|
NM_022062
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr4_-_5890266
|
0.288
|
NM_001313
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr5_+_140186658
|
0.286
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr7_+_89840999
|
0.286
|
NM_001244944
NM_001244945
NM_152999
NM_001040665
NM_001244946
|
STEAP2
|
STEAP family member 2, metalloreductase
|
|
chr3_-_127541975
|
0.284
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr5_+_140247767
|
0.283
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr8_-_70747200
|
0.279
|
NM_030958
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr10_-_21463019
|
0.276
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr12_-_46663207
|
0.271
|
NM_001077484
NM_030674
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr5_+_140753480
|
0.271
|
NM_018919
NM_032086
|
PCDHGA6
|
protocadherin gamma subfamily A, 6
|
|
chr2_-_148779172
|
0.269
|
NM_181742
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr13_-_107187312
|
0.269
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr12_+_111471827
|
0.264
|
NM_015267
|
CUX2
|
cut-like homeobox 2
|
|
chr21_-_38338618
|
0.262
|
NM_001242784
|
HLCS
|
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase)
|
|
chr1_-_37499708
|
0.262
|
NM_000831
|
GRIK3
|
glutamate receptor, ionotropic, kainate 3
|
|
chr9_-_95055989
|
0.261
|
NM_002161
|
IARS
|
isoleucyl-tRNA synthetase
|
|
chr8_-_74884500
|
0.260
|
NM_001204857
NM_001204858
NM_001204859
NM_001204860
NM_001204863
NM_001204864
NM_005648
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C)
|
|
chr12_+_5019048
|
0.258
|
NM_000217
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chr4_-_76439582
|
0.254
|
NM_001008925
NM_001009922
NM_015436
|
RCHY1
|
ring finger and CHY zinc finger domain containing 1
|
|
chr5_+_140739589
|
0.252
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr5_+_139493689
|
0.251
|
NM_005859
|
PURA
|
purine-rich element binding protein A
|
|
chr9_-_98270830
|
0.249
|
NM_000264
|
PTCH1
|
patched 1
|
|
chr5_+_67584251
|
0.245
|
NM_181524
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr1_+_228270360
|
0.244
|
NM_001024226
NM_001658
|
ARF1
|
ADP-ribosylation factor 1
|
|
chr5_+_140710203
|
0.244
|
NM_018912
NM_031993
|
PCDHGA1
|
protocadherin gamma subfamily A, 1
|
|
chr14_+_55518313
|
0.242
|
NM_144578
|
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1-like
|
|
chr16_-_31021716
|
0.240
|
NM_052874
|
STX1B
|
syntaxin 1B
|
|
chr15_-_78423556
|
0.240
|
NM_006383
|
CIB2
|
calcium and integrin binding family member 2
|
|
chr11_-_125933186
|
0.237
|
NM_001243597
NM_016952
|
CDON
|
Cdon homolog (mouse)
|
|
chr5_+_140254930
|
0.236
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr7_+_101460881
|
0.235
|
NM_181552
|
CUX1
|
cut-like homeobox 1
|
|
chrX_+_16804531
|
0.232
|
NM_001168683
NM_018360
|
TXLNG
|
taxilin gamma
|
|
chr1_+_2985730
|
0.232
|
NM_022114
NM_199454
|
PRDM16
|
PR domain containing 16
|
|
chr10_+_89623091
|
0.230
|
NM_000314
|
PTEN
|
phosphatase and tensin homolog
|
|
chr1_+_167189948
|
0.228
|
NM_001198786
NM_002697
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr22_+_40573879
|
0.226
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chrX_+_40944858
|
0.225
|
NM_001039590
NM_001039591
|
USP9X
|
ubiquitin specific peptidase 9, X-linked
|
|
chr3_-_125094003
|
0.223
|
NM_021964
|
ZNF148
|
zinc finger protein 148
|
|
chr9_-_77567735
|
0.223
|
NM_017998
|
C9orf40
|
chromosome 9 open reading frame 40
|