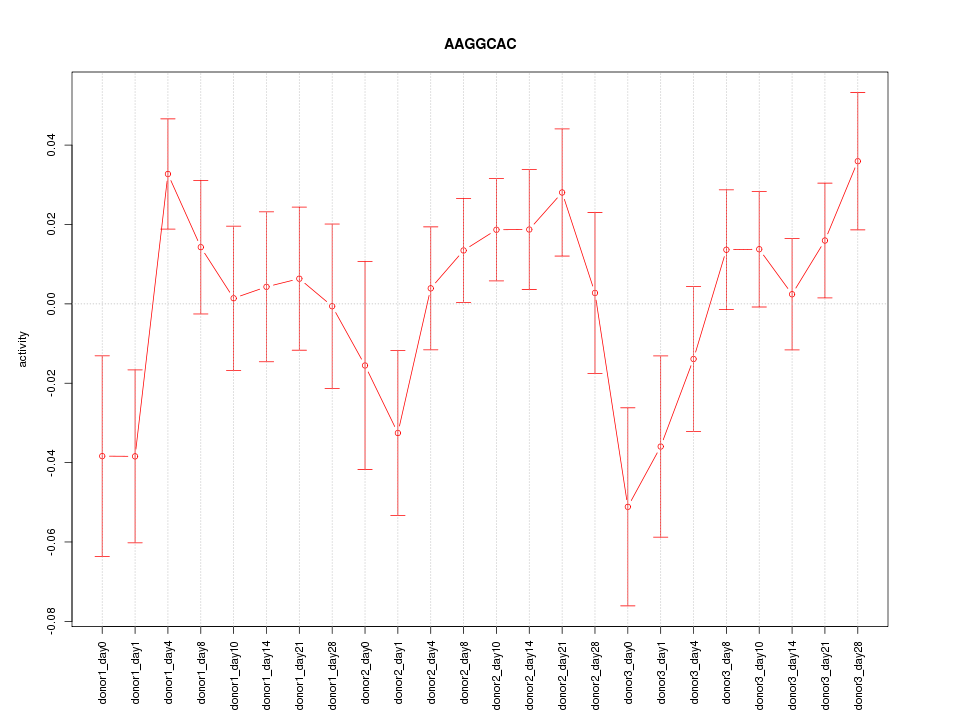
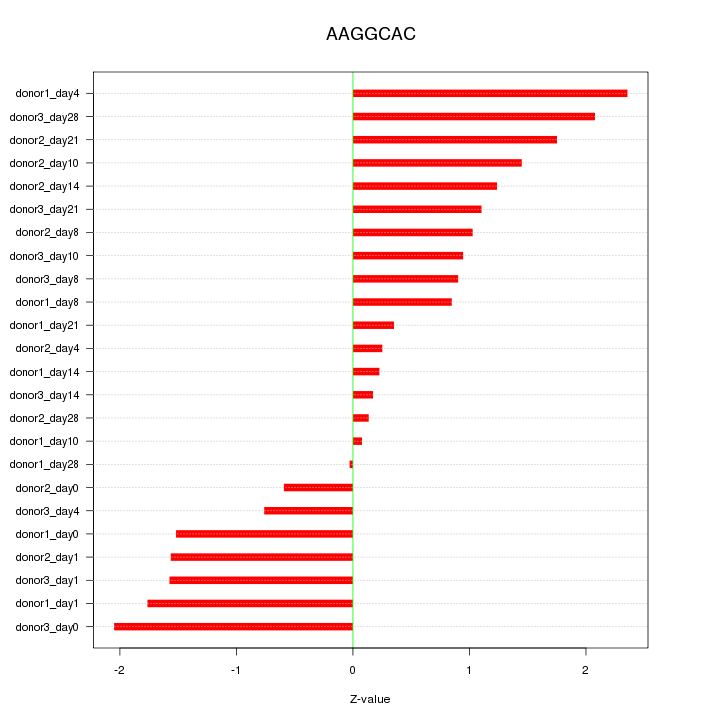
Motif ID: AAGGCAC
Z-value: 1.242
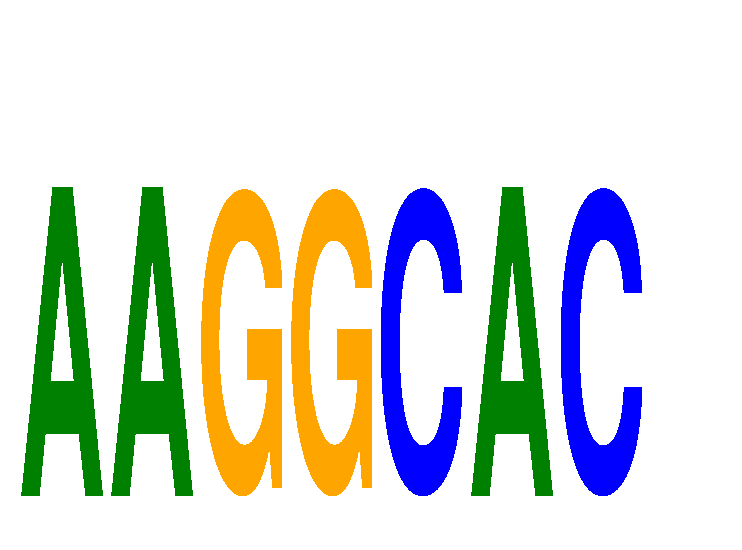
Mature miRNA associated with seed AAGGCAC:
| Name | miRBase Accession |
|---|---|
| hsa-miR-124-3p.1 | MIMAT0000422 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.8 | 2.3 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.6 | 1.8 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.6 | 1.7 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.4 | 1.3 | GO:1903410 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.4 | 2.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.4 | 1.2 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.4 | 0.4 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.4 | 1.2 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.4 | 3.0 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.4 | 0.7 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.4 | 1.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.4 | 1.8 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.4 | 2.9 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.4 | 2.9 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.3 | 2.0 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.3 | 1.0 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.3 | 3.0 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.3 | 0.6 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.3 | 2.2 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.3 | 0.3 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.3 | 1.4 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.3 | 1.8 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.3 | 1.5 | GO:0032803 | regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) |
| 0.2 | 2.3 | GO:0015677 | copper ion import(GO:0015677) |
| 0.2 | 3.3 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.2 | 0.9 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 1.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.2 | 0.9 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.2 | 1.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 0.9 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.2 | 0.7 | GO:1905246 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.2 | 0.9 | GO:0018032 | protein amidation(GO:0018032) |
| 0.2 | 1.3 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.2 | 0.9 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.2 | 1.9 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.2 | 1.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.2 | 1.0 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) positive regulation of NK T cell differentiation(GO:0051138) germline stem cell asymmetric division(GO:0098728) |
| 0.2 | 0.6 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.2 | 0.7 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.2 | 1.1 | GO:1901908 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.2 | 2.5 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.2 | 0.7 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 | 0.5 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.2 | 2.4 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.2 | 0.8 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 | 1.4 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.2 | 2.6 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.1 | 0.4 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.1 | 0.9 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.1 | 2.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.2 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 1.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.4 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.1 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 0.4 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.1 | 1.5 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 0.3 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.1 | 0.7 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.6 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.1 | 1.0 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 1.0 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.4 | GO:0006844 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.1 | 0.5 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 0.9 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.1 | 0.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 1.0 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.1 | 0.3 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 0.3 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.5 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 0.8 | GO:0090032 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 0.1 | 0.2 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 0.7 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 1.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 1.1 | GO:0045964 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.1 | 0.1 | GO:0098907 | regulation of SA node cell action potential(GO:0098907) |
| 0.1 | 1.7 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.5 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 1.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.3 | GO:0035283 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 1.7 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.3 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.1 | 1.3 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.3 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 | 0.4 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 | 1.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.6 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.3 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.7 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 1.0 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.1 | 0.4 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 1.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.8 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 1.5 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.5 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 0.6 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.1 | 0.6 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.7 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) regulation of type B pancreatic cell proliferation(GO:0061469) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.6 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.1 | 1.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.5 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 1.0 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.1 | 1.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 0.3 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.1 | 1.1 | GO:0048863 | stem cell differentiation(GO:0048863) |
| 0.1 | 0.4 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.1 | 1.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.2 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.2 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.1 | 0.2 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.2 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 1.5 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.1 | 1.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 0.6 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.1 | 0.3 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.1 | 0.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.2 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.1 | 0.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.2 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 0.5 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.2 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.1 | 0.1 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 | 0.8 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 1.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 1.1 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.1 | 0.5 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 0.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.2 | GO:0010816 | neuropeptide catabolic process(GO:0010813) substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 2.2 | GO:0060307 | regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) |
| 0.1 | 0.6 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 0.1 | 0.2 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.5 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.3 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.2 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 1.4 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.1 | 1.7 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.1 | 1.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 0.6 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 0.3 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 0.3 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.1 | 1.0 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.2 | GO:0051660 | cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) |
| 0.1 | 1.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.2 | GO:0032455 | nerve growth factor processing(GO:0032455) neurotrophin production(GO:0032898) |
| 0.0 | 1.0 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 2.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0051170 | nuclear import(GO:0051170) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.4 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.1 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.4 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.5 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.4 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.3 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 4.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.5 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.5 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.2 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.4 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 1.1 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.2 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.4 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.3 | GO:0072513 | semicircular canal morphogenesis(GO:0048752) positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 1.8 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.2 | GO:0046676 | negative regulation of peptide secretion(GO:0002792) negative regulation of insulin secretion(GO:0046676) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.0 | 0.2 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.3 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.0 | 0.2 | GO:0034670 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.2 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 | 0.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.8 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 6.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.0 | 0.1 | GO:0035194 | posttranscriptional gene silencing(GO:0016441) posttranscriptional gene silencing by RNA(GO:0035194) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.4 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 1.6 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.2 | GO:0097278 | complement-dependent cytotoxicity(GO:0097278) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.6 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.3 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.0 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.2 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.3 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.0 | 1.3 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.7 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.1 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.7 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.1 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.3 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.3 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 2.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.7 | GO:0060761 | negative regulation of response to cytokine stimulus(GO:0060761) |
| 0.0 | 0.1 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.3 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.5 | GO:0043931 | ossification involved in bone maturation(GO:0043931) |
| 0.0 | 0.2 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.3 | GO:1904262 | TORC2 signaling(GO:0038203) negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.8 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.2 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.4 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 1.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.3 | GO:1903432 | regulation of TORC1 signaling(GO:1903432) |
| 0.0 | 0.3 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) |
| 0.0 | 0.1 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.4 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 0.1 | GO:0051701 | interaction with host(GO:0051701) |
| 0.0 | 0.1 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.3 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.2 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.0 | GO:0090212 | regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.0 | 1.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.9 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.4 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 1.0 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.1 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.0 | 0.4 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 1.3 | GO:0032760 | positive regulation of tumor necrosis factor production(GO:0032760) |
| 0.0 | 1.2 | GO:0030888 | regulation of B cell proliferation(GO:0030888) |
| 0.0 | 0.5 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.3 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.2 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.3 | GO:0034776 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.3 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.4 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.3 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.3 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.0 | 0.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.5 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.8 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.5 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.9 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.8 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.6 | GO:0008593 | regulation of Notch signaling pathway(GO:0008593) |
| 0.0 | 0.1 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.1 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.8 | GO:0061718 | NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.7 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.4 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.0 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.0 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.0 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 0.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 1.1 | GO:0007588 | excretion(GO:0007588) |
| 0.0 | 0.0 | GO:0010560 | positive regulation of glycoprotein biosynthetic process(GO:0010560) |
| 0.0 | 0.0 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.0 | 0.3 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0043507 | positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.2 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.2 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.2 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.3 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.3 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.2 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.2 | GO:0048477 | oogenesis(GO:0048477) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.7 | 2.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.5 | 3.0 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.3 | 0.9 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.3 | 1.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 0.7 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.2 | 2.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.5 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.9 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 2.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.5 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 1.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.5 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.1 | 0.7 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.9 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 1.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 0.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.7 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 1.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.6 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.6 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.5 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.6 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 1.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.2 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 2.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.7 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 0.5 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 0.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.4 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 3.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 1.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.9 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 2.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.4 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.3 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.4 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.1 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 4.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.3 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.2 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.9 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 1.1 | GO:0032991 | macromolecular complex(GO:0032991) |
| 0.0 | 2.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0032279 | axonemal microtubule(GO:0005879) asymmetric synapse(GO:0032279) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.5 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 1.8 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 3.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.4 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.1 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.8 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.1 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.7 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.5 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.4 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 1.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 1.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.4 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 1.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.0 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.6 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.0 | 1.4 | GO:0016363 | nuclear matrix(GO:0016363) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.6 | 2.9 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.4 | 1.7 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.4 | 1.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.4 | 0.4 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.4 | 1.1 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.3 | 1.3 | GO:0005289 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.3 | 1.5 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.3 | 3.0 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.3 | 0.9 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.3 | 1.4 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.3 | 1.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.3 | 2.0 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 0.7 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.2 | 2.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 0.9 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.2 | 1.9 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.2 | 1.0 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.2 | 1.3 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 0.7 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 1.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.2 | 1.1 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.2 | 0.3 | GO:0015184 | sulfur amino acid transmembrane transporter activity(GO:0000099) L-cystine transmembrane transporter activity(GO:0015184) modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.2 | 0.7 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.2 | 0.6 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.2 | 0.5 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.2 | 0.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 0.5 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.2 | 1.8 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.4 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.1 | 0.4 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.4 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.8 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.5 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.1 | 1.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 3.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 0.4 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 2.5 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 0.4 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.4 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 1.8 | GO:0031957 | fatty acid transporter activity(GO:0015245) very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.6 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 0.4 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 1.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.7 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.1 | 1.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.3 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.6 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.5 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.5 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 1.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.1 | 0.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.3 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.3 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.3 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 1.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 2.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.5 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 0.5 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.4 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.7 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 0.7 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 4.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.2 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 1.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.9 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.3 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.1 | 0.9 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.0 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 0.6 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.4 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 0.3 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.1 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 2.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.5 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 2.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 1.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.3 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 0.9 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 1.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.7 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.3 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 1.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.5 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
| 0.0 | 0.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.3 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.9 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 1.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 3.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 1.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.2 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 2.0 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.2 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.3 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 1.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.2 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.3 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 1.9 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA stem-loop binding(GO:0071207) histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 2.8 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 2.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 1.8 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.5 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.5 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 3.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 2.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 1.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 1.1 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.0 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.1 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | PID_EPO_PATHWAY | EPO signaling pathway |
| 0.1 | 1.1 | ST_PAC1_RECEPTOR_PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 1.3 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.0 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 4.0 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.0 | 5.1 | PID_MTOR_4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.6 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.8 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 2.5 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.0 | 1.3 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.2 | PID_MET_PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.4 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.2 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.0 | 1.8 | PID_ERBB4_PATHWAY | ErbB4 signaling events |
| 0.0 | 2.4 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.6 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.4 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.9 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.7 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.1 | PID_CERAMIDE_PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.5 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.2 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.5 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | PID_CD40_PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.8 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.7 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.3 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.0 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.3 | REACTOME_TRAF3_DEPENDENT_IRF_ACTIVATION_PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 2.5 | REACTOME_TRANSPORT_OF_ORGANIC_ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.8 | REACTOME_SIGNALING_BY_EGFR_IN_CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.1 | 0.3 | REACTOME_SHC_MEDIATED_SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 1.5 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.8 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.9 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.1 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.5 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 0.5 | REACTOME_MEMBRANE_BINDING_AND_TARGETTING_OF_GAG_PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.7 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.4 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.4 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 0.6 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.7 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME_PKA_MEDIATED_PHOSPHORYLATION_OF_CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.2 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.6 | REACTOME_REGULATED_PROTEOLYSIS_OF_P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 2.1 | REACTOME_IRON_UPTAKE_AND_TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.7 | REACTOME_ACTIVATED_AMPK_STIMULATES_FATTY_ACID_OXIDATION_IN_MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 5.5 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 3.3 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME_INFLUENZA_VIRAL_RNA_TRANSCRIPTION_AND_REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.4 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.9 | REACTOME_TRAF6_MEDIATED_NFKB_ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 1.0 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.9 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.4 | REACTOME_ROLE_OF_SECOND_MESSENGERS_IN_NETRIN1_SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.7 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME_ORGANIC_CATION_ANION_ZWITTERION_TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 1.7 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.8 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.9 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.3 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME_KERATAN_SULFATE_KERATIN_METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.7 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.5 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME_PPARA_ACTIVATES_GENE_EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.0 | REACTOME_REGULATION_OF_WATER_BALANCE_BY_RENAL_AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 1.5 | REACTOME_TRANS_GOLGI_NETWORK_VESICLE_BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.2 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME_INHIBITION_OF_INSULIN_SECRETION_BY_ADRENALINE_NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.6 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME_SIGNALING_BY_PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.8 | REACTOME_SPHINGOLIPID_METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.4 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME_GENERATION_OF_SECOND_MESSENGER_MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.4 | REACTOME_CONVERSION_FROM_APC_C_CDC20_TO_APC_C_CDH1_IN_LATE_ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME_PRE_NOTCH_EXPRESSION_AND_PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.1 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.4 | REACTOME_SMAD2_SMAD3_SMAD4_HETEROTRIMER_REGULATES_TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |