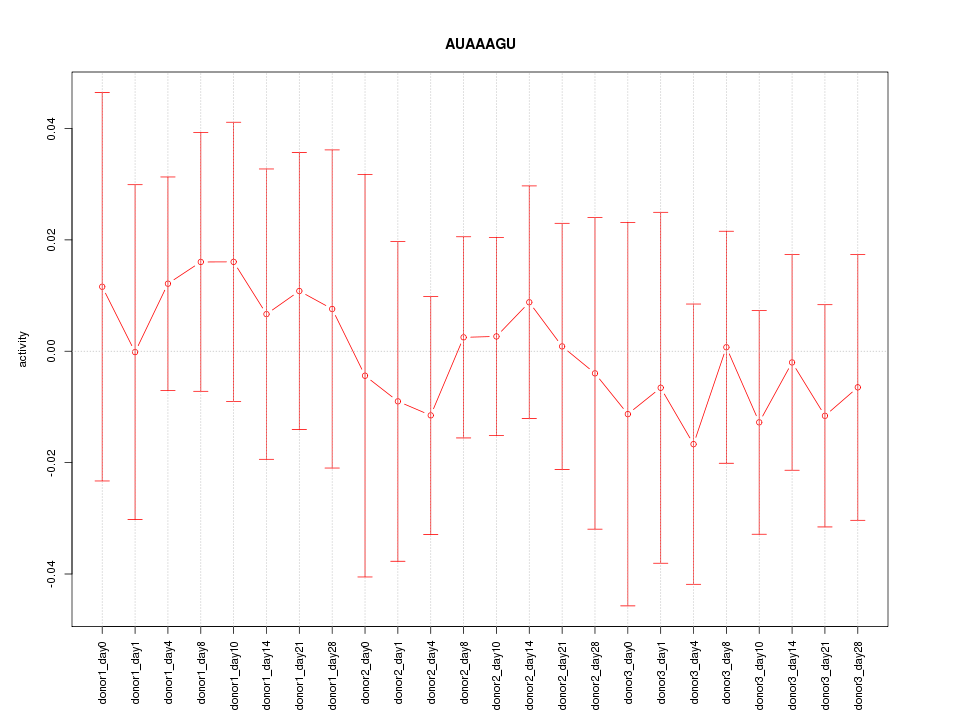
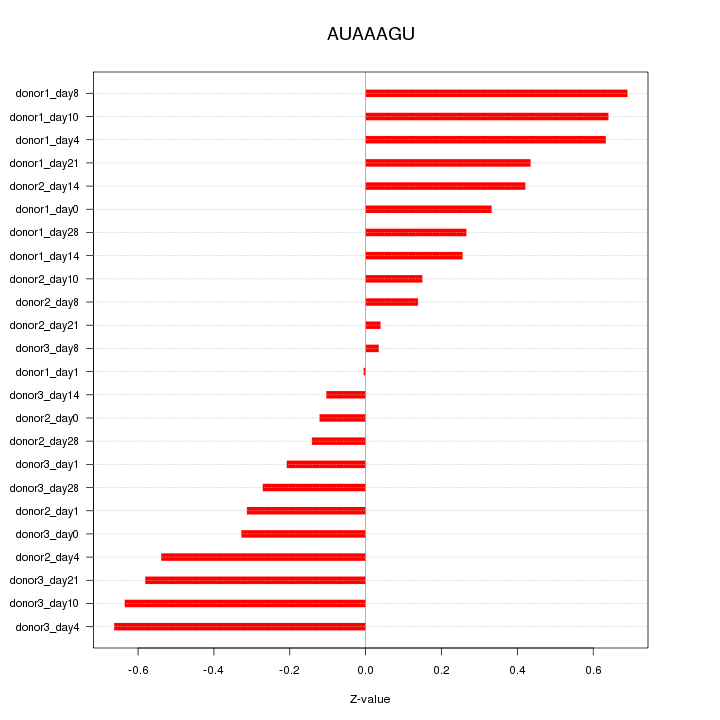
| 0.1 |
0.2 |
GO:0072186 |
metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) |
| 0.1 |
0.3 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 |
0.3 |
GO:0070662 |
mast cell proliferation(GO:0070662) |
| 0.1 |
0.2 |
GO:0015993 |
L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 |
0.5 |
GO:0061470 |
T follicular helper cell differentiation(GO:0061470) |
| 0.1 |
0.2 |
GO:1900169 |
regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 |
0.1 |
GO:0071284 |
cellular response to lead ion(GO:0071284) |
| 0.0 |
0.1 |
GO:0097026 |
dendritic cell dendrite assembly(GO:0097026) |
| 0.0 |
0.1 |
GO:1902725 |
negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 |
0.2 |
GO:0060235 |
lens induction in camera-type eye(GO:0060235) |
| 0.0 |
0.1 |
GO:0006288 |
base-excision repair, DNA ligation(GO:0006288) |
| 0.0 |
0.1 |
GO:0034059 |
response to anoxia(GO:0034059) |
| 0.0 |
0.1 |
GO:0071879 |
positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 |
0.1 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 |
0.1 |
GO:0035936 |
testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.0 |
0.1 |
GO:0060490 |
orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 |
0.1 |
GO:2000546 |
positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 |
0.1 |
GO:0015798 |
myo-inositol transport(GO:0015798) |
| 0.0 |
0.1 |
GO:1900224 |
positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 |
0.1 |
GO:0033387 |
putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 |
0.0 |
GO:0060775 |
mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.0 |
0.0 |
GO:0021823 |
cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 |
0.2 |
GO:0071169 |
establishment of protein localization to chromatin(GO:0071169) |
| 0.0 |
0.1 |
GO:0060398 |
regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 |
0.0 |
GO:2000705 |
dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 |
0.1 |
GO:0048382 |
lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) mesendoderm development(GO:0048382) |
| 0.0 |
0.2 |
GO:0045054 |
constitutive secretory pathway(GO:0045054) |
| 0.0 |
0.0 |
GO:0034140 |
negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 |
0.1 |
GO:0046952 |
ketone body catabolic process(GO:0046952) |
| 0.0 |
0.2 |
GO:0033601 |
positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 |
0.0 |
GO:2000118 |
regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 |
0.0 |
GO:0003162 |
atrioventricular node development(GO:0003162) |
| 0.0 |
0.0 |
GO:1904395 |
positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 |
0.1 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.0 |
0.2 |
GO:0001886 |
endothelial cell morphogenesis(GO:0001886) |
| 0.0 |
0.3 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.0 |
0.1 |
GO:2000645 |
negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 |
0.1 |
GO:0000710 |
meiotic mismatch repair(GO:0000710) |
| 0.0 |
0.2 |
GO:0010944 |
negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 |
0.1 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.0 |
0.0 |
GO:1903984 |
negative regulation of ribosome biogenesis(GO:0090071) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 |
0.0 |
GO:0051987 |
positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 |
0.1 |
GO:0010609 |
mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.0 |
0.1 |
GO:0008343 |
adult feeding behavior(GO:0008343) |
| 0.0 |
0.1 |
GO:0071233 |
cellular response to leucine(GO:0071233) |
| 0.0 |
0.1 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.0 |
0.1 |
GO:0060437 |
lung growth(GO:0060437) |
| 0.0 |
0.0 |
GO:0031548 |
regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 |
0.0 |
GO:0071596 |
ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 |
0.1 |
GO:0030047 |
actin modification(GO:0030047) |