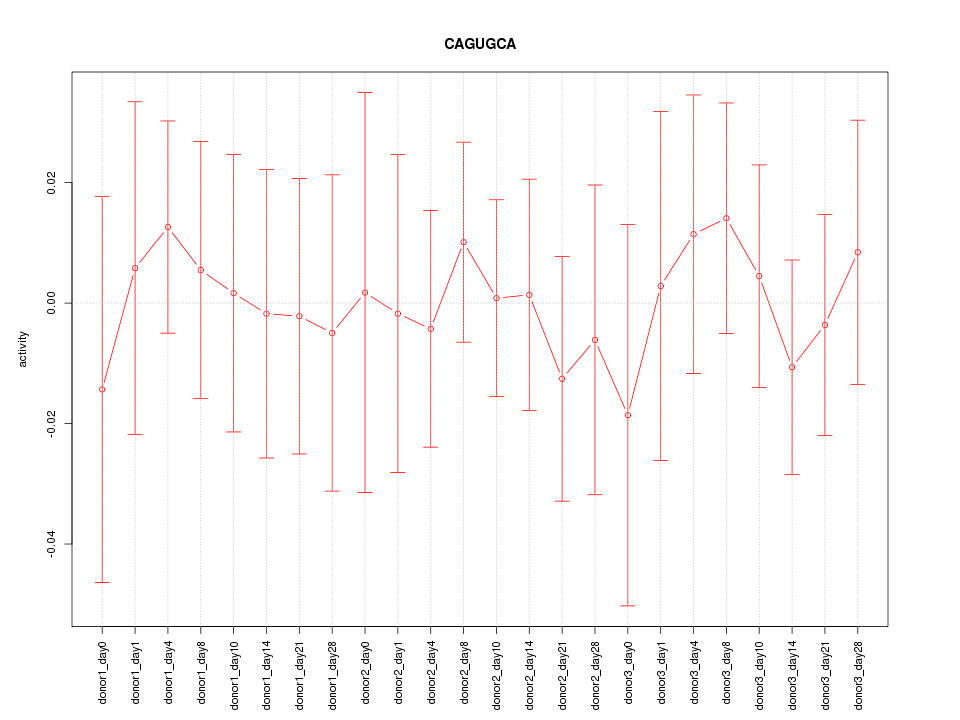
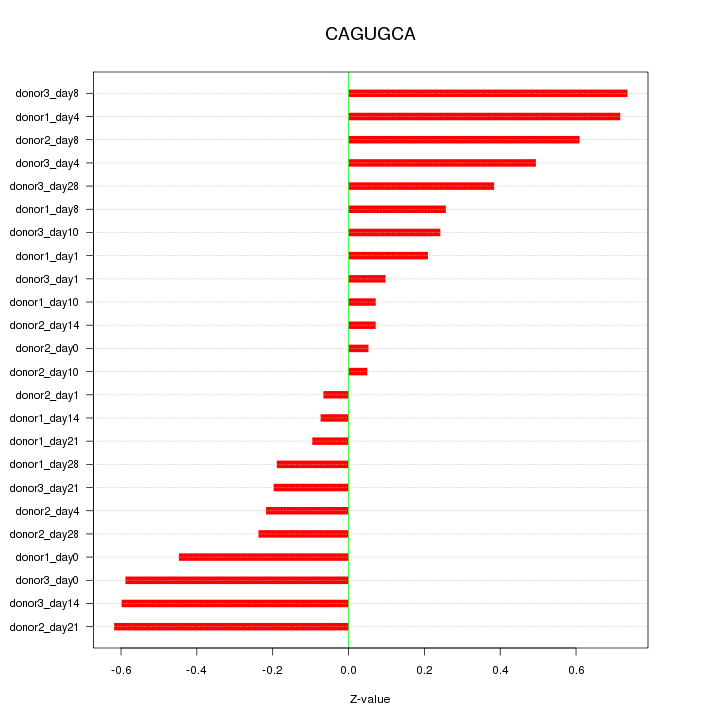
| 0.1 |
0.3 |
GO:1902256 |
apoptotic process involved in outflow tract morphogenesis(GO:0003275) negative regulation of alkaline phosphatase activity(GO:0010693) uterine wall breakdown(GO:0042704) positive regulation of catagen(GO:0051795) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.1 |
0.2 |
GO:1901860 |
positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.1 |
0.2 |
GO:0035283 |
rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 |
0.3 |
GO:0051138 |
positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 |
0.2 |
GO:0014740 |
negative regulation of muscle hyperplasia(GO:0014740) |
| 0.0 |
0.1 |
GO:0021823 |
cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 |
0.2 |
GO:0032792 |
negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 |
0.0 |
GO:0003032 |
detection of oxygen(GO:0003032) |
| 0.0 |
0.2 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 |
0.1 |
GO:0033634 |
positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 |
0.1 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 |
0.4 |
GO:0060628 |
regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 |
0.1 |
GO:0032877 |
positive regulation of DNA endoreduplication(GO:0032877) negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 |
0.1 |
GO:2000097 |
chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 |
0.1 |
GO:0006679 |
glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 |
0.1 |
GO:0007206 |
phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 |
0.1 |
GO:0060708 |
spongiotrophoblast differentiation(GO:0060708) |
| 0.0 |
0.1 |
GO:0098907 |
protein localization to M-band(GO:0036309) regulation of SA node cell action potential(GO:0098907) |
| 0.0 |
0.0 |
GO:0061428 |
negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 |
0.2 |
GO:0014807 |
regulation of somitogenesis(GO:0014807) |
| 0.0 |
0.1 |
GO:0043129 |
surfactant homeostasis(GO:0043129) |
| 0.0 |
0.1 |
GO:1905098 |
negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 |
0.1 |
GO:0015917 |
aminophospholipid transport(GO:0015917) regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 |
0.0 |
GO:0045925 |
positive regulation of female receptivity(GO:0045925) |
| 0.0 |
0.0 |
GO:0003241 |
growth involved in heart morphogenesis(GO:0003241) |
| 0.0 |
0.1 |
GO:0051987 |
positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 |
0.1 |
GO:0010890 |
positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 |
0.1 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.1 |
GO:0098967 |
exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 |
0.0 |
GO:0033242 |
regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 |
0.0 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.0 |
0.1 |
GO:0070649 |
polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 |
0.0 |
GO:0052151 |
positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 |
0.0 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 |
0.1 |
GO:2000580 |
positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 |
0.3 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.0 |
0.0 |
GO:1903526 |
negative regulation of membrane tubulation(GO:1903526) |