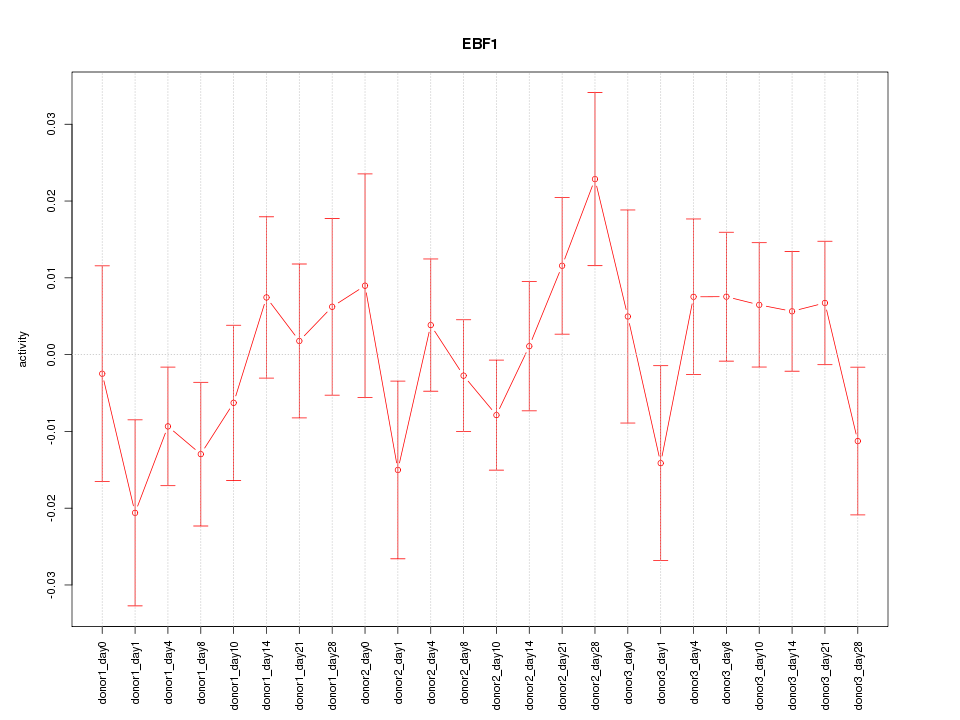
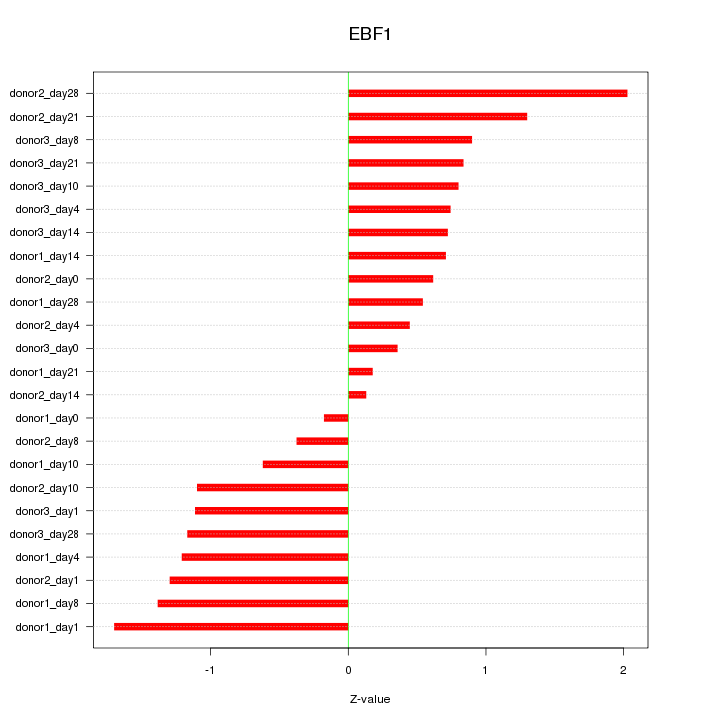
| 0.8 |
3.9 |
GO:0060741 |
prostate gland stromal morphogenesis(GO:0060741) |
| 0.4 |
1.1 |
GO:1904395 |
positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.3 |
1.0 |
GO:0006667 |
sphinganine metabolic process(GO:0006667) |
| 0.3 |
0.3 |
GO:0018199 |
peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) peptidyl-glutamine modification(GO:0018199) |
| 0.3 |
1.7 |
GO:1990822 |
basic amino acid transmembrane transport(GO:1990822) |
| 0.3 |
1.4 |
GO:0072313 |
metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.2 |
1.0 |
GO:0072658 |
maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 |
1.2 |
GO:0002901 |
mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.2 |
1.5 |
GO:0010734 |
protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.2 |
0.2 |
GO:0034447 |
very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.2 |
1.2 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.2 |
0.7 |
GO:0043602 |
nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) |
| 0.2 |
1.3 |
GO:0035469 |
determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.2 |
1.4 |
GO:0045007 |
depurination(GO:0045007) |
| 0.2 |
0.5 |
GO:0006566 |
threonine metabolic process(GO:0006566) |
| 0.2 |
0.7 |
GO:0002086 |
diaphragm contraction(GO:0002086) |
| 0.2 |
0.8 |
GO:1904158 |
axonemal central apparatus assembly(GO:1904158) |
| 0.2 |
0.5 |
GO:1905072 |
apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.2 |
0.3 |
GO:0060164 |
regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 |
0.1 |
GO:0060433 |
bronchus development(GO:0060433) |
| 0.1 |
0.4 |
GO:0014016 |
neuroblast differentiation(GO:0014016) |
| 0.1 |
0.7 |
GO:0002378 |
immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 |
0.4 |
GO:0090675 |
intermicrovillar adhesion(GO:0090675) |
| 0.1 |
0.4 |
GO:0038193 |
thromboxane A2 signaling pathway(GO:0038193) |
| 0.1 |
0.4 |
GO:0030186 |
melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 |
0.3 |
GO:0001560 |
regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 |
0.2 |
GO:0042323 |
negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.1 |
0.1 |
GO:0060037 |
pharyngeal system development(GO:0060037) |
| 0.1 |
0.4 |
GO:0033277 |
abortive mitotic cell cycle(GO:0033277) |
| 0.1 |
0.3 |
GO:0034239 |
macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.1 |
0.3 |
GO:0050760 |
negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 |
0.4 |
GO:2001295 |
malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 |
0.3 |
GO:2000661 |
positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.1 |
0.5 |
GO:0050917 |
sensory perception of umami taste(GO:0050917) |
| 0.1 |
0.3 |
GO:0090310 |
negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 |
0.3 |
GO:0016185 |
synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 |
0.9 |
GO:0006477 |
protein sulfation(GO:0006477) |
| 0.1 |
0.1 |
GO:0036309 |
protein localization to M-band(GO:0036309) |
| 0.1 |
0.6 |
GO:0098704 |
fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.1 |
0.3 |
GO:0060545 |
CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) positive regulation of necroptotic process(GO:0060545) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.1 |
0.3 |
GO:0055073 |
cadmium ion homeostasis(GO:0055073) divalent metal ion export(GO:0070839) |
| 0.1 |
0.6 |
GO:0050915 |
sensory perception of sour taste(GO:0050915) |
| 0.1 |
0.3 |
GO:1905246 |
regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 |
0.1 |
GO:0090024 |
negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 |
0.3 |
GO:1900111 |
positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.1 |
0.2 |
GO:1904604 |
regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904596) negative regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904597) regulation of advanced glycation end-product receptor activity(GO:1904603) negative regulation of advanced glycation end-product receptor activity(GO:1904604) negative regulation of connective tissue replacement(GO:1905204) |
| 0.1 |
0.7 |
GO:0018916 |
nitrobenzene metabolic process(GO:0018916) |
| 0.1 |
0.4 |
GO:0060935 |
cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 0.1 |
1.0 |
GO:0008343 |
adult feeding behavior(GO:0008343) |
| 0.1 |
0.2 |
GO:1901052 |
sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.1 |
0.4 |
GO:0032474 |
otolith morphogenesis(GO:0032474) sensory system development(GO:0048880) |
| 0.1 |
0.3 |
GO:1902613 |
regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.1 |
0.5 |
GO:0001712 |
ectodermal cell fate commitment(GO:0001712) |
| 0.1 |
0.3 |
GO:0006210 |
pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 |
0.6 |
GO:0061669 |
spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 |
0.2 |
GO:0090294 |
nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.1 |
0.2 |
GO:0060708 |
spongiotrophoblast differentiation(GO:0060708) |
| 0.1 |
0.2 |
GO:2000824 |
negative regulation of androgen receptor activity(GO:2000824) |
| 0.1 |
0.4 |
GO:0002925 |
positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 |
0.7 |
GO:0051388 |
positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 |
0.2 |
GO:0006174 |
dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.1 |
0.3 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.1 |
1.0 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.1 |
0.2 |
GO:0034146 |
toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.1 |
0.3 |
GO:1904491 |
protein localization to ciliary transition zone(GO:1904491) |
| 0.1 |
0.2 |
GO:0071109 |
superior temporal gyrus development(GO:0071109) |
| 0.1 |
0.2 |
GO:0099545 |
trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 |
0.3 |
GO:0048861 |
leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 |
0.1 |
GO:0003129 |
heart induction(GO:0003129) negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) cardiac cell fate determination(GO:0060913) |
| 0.1 |
1.7 |
GO:0006068 |
ethanol catabolic process(GO:0006068) |
| 0.1 |
0.2 |
GO:0061188 |
negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 |
0.2 |
GO:0060086 |
circadian temperature homeostasis(GO:0060086) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.1 |
0.3 |
GO:1900168 |
negative regulation of circadian sleep/wake cycle, sleep(GO:0042321) glial cell-derived neurotrophic factor secretion(GO:0044467) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.1 |
0.2 |
GO:1903691 |
positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 |
0.9 |
GO:0042048 |
olfactory behavior(GO:0042048) |
| 0.1 |
0.1 |
GO:1903779 |
regulation of cardiac conduction(GO:1903779) |
| 0.1 |
0.2 |
GO:0033686 |
positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.1 |
0.2 |
GO:0002071 |
glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.1 |
1.2 |
GO:0009437 |
carnitine metabolic process(GO:0009437) |
| 0.1 |
0.2 |
GO:0032764 |
negative regulation of mast cell cytokine production(GO:0032764) |
| 0.1 |
1.8 |
GO:0044458 |
motile cilium assembly(GO:0044458) |
| 0.1 |
0.1 |
GO:0061010 |
gall bladder development(GO:0061010) |
| 0.1 |
0.3 |
GO:0030241 |
skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 |
0.2 |
GO:0001798 |
positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 |
0.1 |
GO:0031630 |
regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.1 |
0.2 |
GO:0038178 |
complement component C5a signaling pathway(GO:0038178) |
| 0.1 |
0.2 |
GO:0090156 |
cellular sphingolipid homeostasis(GO:0090156) |
| 0.1 |
0.2 |
GO:0090271 |
positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 |
0.2 |
GO:0018057 |
peptidyl-lysine oxidation(GO:0018057) |
| 0.1 |
0.2 |
GO:0072156 |
distal tubule morphogenesis(GO:0072156) |
| 0.1 |
0.2 |
GO:1902995 |
regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.1 |
0.2 |
GO:0005988 |
lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.1 |
0.4 |
GO:0070309 |
lens fiber cell morphogenesis(GO:0070309) |
| 0.1 |
0.3 |
GO:0070383 |
DNA cytosine deamination(GO:0070383) |
| 0.1 |
0.2 |
GO:0021553 |
olfactory nerve development(GO:0021553) |
| 0.1 |
0.4 |
GO:0033590 |
response to cobalamin(GO:0033590) |
| 0.1 |
0.1 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) |
| 0.1 |
0.2 |
GO:0072229 |
proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) metanephric proximal tubule development(GO:0072237) |
| 0.1 |
0.2 |
GO:1901842 |
negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 |
0.3 |
GO:1900920 |
regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.1 |
0.3 |
GO:0070560 |
protein secretion by platelet(GO:0070560) |
| 0.1 |
0.4 |
GO:0070779 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 |
0.2 |
GO:0002644 |
negative regulation of tolerance induction(GO:0002644) |
| 0.1 |
0.2 |
GO:2000721 |
positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.1 |
0.4 |
GO:0070235 |
regulation of activation-induced cell death of T cells(GO:0070235) |
| 0.1 |
0.5 |
GO:2001300 |
lipoxin metabolic process(GO:2001300) |
| 0.1 |
0.2 |
GO:2001190 |
positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 |
0.2 |
GO:0070253 |
somatostatin secretion(GO:0070253) |
| 0.1 |
0.2 |
GO:0035494 |
SNARE complex disassembly(GO:0035494) |
| 0.1 |
0.1 |
GO:1901963 |
regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) |
| 0.1 |
0.3 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.1 |
0.4 |
GO:0070649 |
polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 |
0.2 |
GO:0006740 |
NADPH regeneration(GO:0006740) |
| 0.1 |
0.9 |
GO:0060850 |
regulation of transcription involved in cell fate commitment(GO:0060850) |
| 0.1 |
0.2 |
GO:0060120 |
auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 |
0.3 |
GO:0032729 |
positive regulation of interferon-gamma production(GO:0032729) |
| 0.0 |
0.3 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 |
0.3 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) |
| 0.0 |
0.1 |
GO:0060994 |
regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.0 |
0.2 |
GO:0072092 |
ureteric bud invasion(GO:0072092) |
| 0.0 |
0.0 |
GO:0003160 |
endocardium morphogenesis(GO:0003160) |
| 0.0 |
0.3 |
GO:0006196 |
AMP catabolic process(GO:0006196) |
| 0.0 |
0.5 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 |
0.1 |
GO:0044861 |
protein transport into plasma membrane raft(GO:0044861) |
| 0.0 |
0.1 |
GO:2000977 |
regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) regulation of forebrain neuron differentiation(GO:2000977) |
| 0.0 |
1.1 |
GO:0016254 |
preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 |
0.2 |
GO:1904479 |
negative regulation of intestinal absorption(GO:1904479) |
| 0.0 |
0.1 |
GO:0090291 |
negative regulation of osteoclast proliferation(GO:0090291) |
| 0.0 |
0.0 |
GO:0071910 |
determination of liver left/right asymmetry(GO:0071910) |
| 0.0 |
0.8 |
GO:0003084 |
positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 |
0.2 |
GO:0000957 |
mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 |
0.0 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 |
0.1 |
GO:0071106 |
coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.0 |
1.4 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.0 |
0.6 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
0.0 |
GO:0038003 |
opioid receptor signaling pathway(GO:0038003) |
| 0.0 |
0.3 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.0 |
0.2 |
GO:0032229 |
negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 |
0.3 |
GO:2000834 |
androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 0.0 |
0.2 |
GO:1900245 |
positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 |
0.1 |
GO:0097252 |
oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 |
0.0 |
GO:0015838 |
quaternary ammonium group transport(GO:0015697) amino-acid betaine transport(GO:0015838) carnitine transport(GO:0015879) |
| 0.0 |
0.3 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 |
0.2 |
GO:1902075 |
cellular response to salt(GO:1902075) |
| 0.0 |
0.1 |
GO:0043385 |
mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.0 |
0.7 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.3 |
GO:2000503 |
positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 |
0.2 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
1.0 |
GO:0055075 |
potassium ion homeostasis(GO:0055075) |
| 0.0 |
0.4 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 |
0.4 |
GO:0031444 |
slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 |
0.4 |
GO:0051902 |
negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 |
0.4 |
GO:0043152 |
induction of bacterial agglutination(GO:0043152) |
| 0.0 |
0.4 |
GO:1901223 |
negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 |
0.1 |
GO:0038163 |
thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 |
0.2 |
GO:1903546 |
protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 |
0.3 |
GO:0098887 |
neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 |
0.2 |
GO:0086046 |
membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 |
0.2 |
GO:0031338 |
regulation of vesicle fusion(GO:0031338) |
| 0.0 |
0.1 |
GO:0080154 |
regulation of fertilization(GO:0080154) |
| 0.0 |
0.9 |
GO:0060716 |
labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 |
0.1 |
GO:0070105 |
positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 |
0.2 |
GO:0043366 |
beta selection(GO:0043366) |
| 0.0 |
0.2 |
GO:0034154 |
toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 |
0.2 |
GO:0006789 |
bilirubin conjugation(GO:0006789) |
| 0.0 |
0.2 |
GO:1903553 |
positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 |
0.1 |
GO:1903697 |
negative regulation of microvillus assembly(GO:1903697) |
| 0.0 |
0.1 |
GO:0070676 |
intralumenal vesicle formation(GO:0070676) |
| 0.0 |
0.3 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 |
0.0 |
GO:0097104 |
postsynaptic membrane assembly(GO:0097104) positive regulation of presynaptic membrane organization(GO:1901631) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 |
0.4 |
GO:2000304 |
positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 |
0.1 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 |
0.1 |
GO:0006624 |
vacuolar protein processing(GO:0006624) |
| 0.0 |
0.2 |
GO:0045084 |
positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 |
0.4 |
GO:0039536 |
negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 |
0.2 |
GO:0046968 |
peptide antigen transport(GO:0046968) |
| 0.0 |
0.4 |
GO:0097500 |
receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 |
0.5 |
GO:0070050 |
neuron cellular homeostasis(GO:0070050) |
| 0.0 |
0.1 |
GO:0098886 |
modification of dendritic spine(GO:0098886) |
| 0.0 |
0.3 |
GO:0070544 |
histone H3-K36 demethylation(GO:0070544) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 |
1.1 |
GO:0035338 |
long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 |
0.2 |
GO:0033211 |
adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 |
0.1 |
GO:0046056 |
dADP metabolic process(GO:0046056) |
| 0.0 |
0.0 |
GO:0061014 |
positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 |
0.3 |
GO:0032776 |
DNA methylation on cytosine(GO:0032776) |
| 0.0 |
0.1 |
GO:0071907 |
determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 |
0.2 |
GO:1901090 |
regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.0 |
0.1 |
GO:1901895 |
negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 |
0.4 |
GO:0014722 |
regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 |
0.2 |
GO:0006651 |
diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 |
0.2 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 |
0.1 |
GO:2001178 |
mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.0 |
0.6 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.1 |
GO:0007512 |
adult heart development(GO:0007512) |
| 0.0 |
0.1 |
GO:0009386 |
translational attenuation(GO:0009386) |
| 0.0 |
0.4 |
GO:0090292 |
nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 |
0.6 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.0 |
0.2 |
GO:0018094 |
protein polyglycylation(GO:0018094) |
| 0.0 |
0.1 |
GO:1902080 |
regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.0 |
0.2 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.0 |
0.0 |
GO:0042346 |
positive regulation of NF-kappaB import into nucleus(GO:0042346) |
| 0.0 |
0.2 |
GO:0034227 |
tRNA thio-modification(GO:0034227) |
| 0.0 |
0.3 |
GO:0001675 |
acrosome assembly(GO:0001675) |
| 0.0 |
0.5 |
GO:0007196 |
adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 |
0.5 |
GO:0015812 |
gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 |
0.3 |
GO:2000270 |
negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 |
0.5 |
GO:0060080 |
inhibitory postsynaptic potential(GO:0060080) |
| 0.0 |
0.2 |
GO:0070445 |
oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 |
0.1 |
GO:0070294 |
renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 |
0.2 |
GO:1904823 |
pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 |
0.1 |
GO:0001807 |
type IV hypersensitivity(GO:0001806) regulation of type IV hypersensitivity(GO:0001807) negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 |
1.2 |
GO:0035082 |
axoneme assembly(GO:0035082) |
| 0.0 |
0.2 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 |
0.2 |
GO:0051140 |
regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 |
0.3 |
GO:0070257 |
positive regulation of mucus secretion(GO:0070257) |
| 0.0 |
0.1 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 |
0.1 |
GO:0061428 |
negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 |
0.0 |
GO:0045168 |
cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 0.0 |
0.2 |
GO:0061055 |
myotome development(GO:0061055) |
| 0.0 |
0.1 |
GO:0072683 |
T cell extravasation(GO:0072683) |
| 0.0 |
0.1 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 |
0.1 |
GO:1903348 |
positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 |
0.2 |
GO:0035021 |
negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 |
0.1 |
GO:0060392 |
negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 |
0.5 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 |
0.4 |
GO:2000651 |
positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 |
0.5 |
GO:1901750 |
leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 |
0.1 |
GO:0043095 |
regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 |
0.1 |
GO:0007406 |
negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 |
0.1 |
GO:0048213 |
Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 |
0.3 |
GO:0006600 |
creatine metabolic process(GO:0006600) |
| 0.0 |
0.1 |
GO:0071638 |
negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 |
0.1 |
GO:0031587 |
positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 |
0.1 |
GO:0046835 |
carbohydrate phosphorylation(GO:0046835) |
| 0.0 |
1.2 |
GO:0048741 |
skeletal muscle fiber development(GO:0048741) |
| 0.0 |
0.1 |
GO:1902723 |
negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 |
0.1 |
GO:0042822 |
pyridoxal phosphate metabolic process(GO:0042822) |
| 0.0 |
0.4 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 |
0.3 |
GO:0043651 |
linoleic acid metabolic process(GO:0043651) |
| 0.0 |
0.3 |
GO:0033008 |
positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 |
0.3 |
GO:0010459 |
negative regulation of heart rate(GO:0010459) |
| 0.0 |
0.1 |
GO:0003050 |
regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 |
0.1 |
GO:0010265 |
SCF complex assembly(GO:0010265) |
| 0.0 |
0.5 |
GO:0071786 |
endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 |
0.2 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.0 |
0.3 |
GO:0002480 |
antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 |
0.1 |
GO:0044313 |
protein K6-linked deubiquitination(GO:0044313) |
| 0.0 |
0.5 |
GO:1904714 |
regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 |
0.5 |
GO:0035640 |
exploration behavior(GO:0035640) |
| 0.0 |
0.1 |
GO:0060440 |
trachea formation(GO:0060440) |
| 0.0 |
0.2 |
GO:1903659 |
regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 |
0.1 |
GO:0035621 |
ER to Golgi ceramide transport(GO:0035621) |
| 0.0 |
0.1 |
GO:0071469 |
sensory perception of touch(GO:0050975) detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.0 |
1.0 |
GO:0050912 |
detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.0 |
0.2 |
GO:0032286 |
central nervous system myelin maintenance(GO:0032286) |
| 0.0 |
0.1 |
GO:0014009 |
glial cell proliferation(GO:0014009) |
| 0.0 |
0.0 |
GO:0090427 |
activation of meiosis(GO:0090427) |
| 0.0 |
0.5 |
GO:0032098 |
regulation of appetite(GO:0032098) |
| 0.0 |
0.2 |
GO:1902219 |
negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 |
0.1 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 |
0.0 |
GO:0045626 |
negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.0 |
0.1 |
GO:0048511 |
rhythmic process(GO:0048511) |
| 0.0 |
0.2 |
GO:0048105 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.0 |
0.0 |
GO:1903281 |
regulation of the force of heart contraction by cardiac conduction(GO:0086092) regulation of calcium:sodium antiporter activity(GO:1903279) positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.0 |
0.1 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 |
0.1 |
GO:0014848 |
urinary bladder smooth muscle contraction(GO:0014832) urinary tract smooth muscle contraction(GO:0014848) |
| 0.0 |
0.1 |
GO:0010730 |
negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 |
0.1 |
GO:0090043 |
regulation of tubulin deacetylation(GO:0090043) |
| 0.0 |
0.2 |
GO:0072592 |
oxygen metabolic process(GO:0072592) |
| 0.0 |
0.1 |
GO:1902093 |
positive regulation of sperm motility(GO:1902093) |
| 0.0 |
0.0 |
GO:0070244 |
negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.0 |
0.1 |
GO:1902361 |
mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 |
0.3 |
GO:1902260 |
negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 |
0.1 |
GO:0034443 |
regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 |
0.1 |
GO:0030046 |
parallel actin filament bundle assembly(GO:0030046) |
| 0.0 |
0.9 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.0 |
0.1 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 |
0.4 |
GO:0010745 |
negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 |
0.1 |
GO:0060385 |
axonogenesis involved in innervation(GO:0060385) |
| 0.0 |
0.2 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.1 |
GO:1900276 |
regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 |
0.0 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.0 |
2.4 |
GO:0006501 |
C-terminal protein lipidation(GO:0006501) |
| 0.0 |
1.0 |
GO:0007340 |
acrosome reaction(GO:0007340) |
| 0.0 |
0.2 |
GO:0060056 |
mammary gland involution(GO:0060056) |
| 0.0 |
0.1 |
GO:1903597 |
negative regulation of gap junction assembly(GO:1903597) |
| 0.0 |
0.4 |
GO:0010603 |
regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 |
0.1 |
GO:0045872 |
positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 |
0.2 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.0 |
0.1 |
GO:1904578 |
response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) response to hypobaric hypoxia(GO:1990910) |
| 0.0 |
0.1 |
GO:0044828 |
negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 |
0.0 |
GO:0046122 |
purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 |
0.2 |
GO:0008286 |
insulin receptor signaling pathway(GO:0008286) |
| 0.0 |
0.2 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 |
0.4 |
GO:1990845 |
adaptive thermogenesis(GO:1990845) |
| 0.0 |
0.1 |
GO:0009447 |
putrescine catabolic process(GO:0009447) |
| 0.0 |
0.2 |
GO:0006572 |
tyrosine catabolic process(GO:0006572) |
| 0.0 |
0.1 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 |
0.7 |
GO:0022400 |
regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 |
0.2 |
GO:0050847 |
progesterone receptor signaling pathway(GO:0050847) |
| 0.0 |
0.0 |
GO:0034465 |
response to carbon monoxide(GO:0034465) |
| 0.0 |
0.1 |
GO:0042560 |
folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 |
0.1 |
GO:0043932 |
ossification involved in bone remodeling(GO:0043932) |
| 0.0 |
0.2 |
GO:1902659 |
regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 |
0.2 |
GO:0001865 |
NK T cell differentiation(GO:0001865) |
| 0.0 |
0.5 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.4 |
GO:0032703 |
negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 |
0.2 |
GO:0060453 |
regulation of gastric acid secretion(GO:0060453) |
| 0.0 |
0.1 |
GO:0072553 |
terminal button organization(GO:0072553) |
| 0.0 |
0.0 |
GO:1904892 |
regulation of JAK-STAT cascade(GO:0046425) regulation of STAT cascade(GO:1904892) |
| 0.0 |
0.2 |
GO:0046465 |
dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.0 |
0.2 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.0 |
0.2 |
GO:1901525 |
negative regulation of macromitophagy(GO:1901525) |
| 0.0 |
0.1 |
GO:1902748 |
positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 |
0.2 |
GO:0007598 |
blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 |
0.5 |
GO:0060122 |
inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 |
0.1 |
GO:0042489 |
negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.0 |
0.1 |
GO:0098707 |
ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 |
0.0 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.0 |
0.1 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.0 |
0.1 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 |
0.2 |
GO:0014041 |
regulation of neuron maturation(GO:0014041) |
| 0.0 |
0.1 |
GO:0033274 |
response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 |
0.1 |
GO:0042421 |
norepinephrine biosynthetic process(GO:0042421) |
| 0.0 |
0.1 |
GO:1905224 |
clathrin-coated pit assembly(GO:1905224) |
| 0.0 |
0.3 |
GO:0021794 |
thalamus development(GO:0021794) |
| 0.0 |
0.2 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 |
0.2 |
GO:0032460 |
negative regulation of protein oligomerization(GO:0032460) |
| 0.0 |
0.1 |
GO:0050884 |
neuromuscular process controlling posture(GO:0050884) |
| 0.0 |
0.1 |
GO:0048250 |
mitochondrial iron ion transport(GO:0048250) |
| 0.0 |
0.2 |
GO:0042339 |
keratan sulfate biosynthetic process(GO:0018146) keratan sulfate metabolic process(GO:0042339) |
| 0.0 |
0.1 |
GO:0002115 |
store-operated calcium entry(GO:0002115) |
| 0.0 |
0.1 |
GO:2000395 |
regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 |
0.0 |
GO:0045226 |
extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 |
0.1 |
GO:1904721 |
negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 |
0.0 |
GO:1902567 |
negative regulation of eosinophil activation(GO:1902567) |
| 0.0 |
0.1 |
GO:0033578 |
protein glycosylation in Golgi(GO:0033578) |
| 0.0 |
0.1 |
GO:0038060 |
nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 |
0.0 |
GO:0060842 |
arterial endothelial cell differentiation(GO:0060842) |
| 0.0 |
0.2 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 |
0.0 |
GO:0061526 |
acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) acetylcholine secretion(GO:0061526) |
| 0.0 |
0.1 |
GO:2000612 |
thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 |
0.0 |
GO:0030222 |
eosinophil differentiation(GO:0030222) |
| 0.0 |
0.3 |
GO:0043923 |
positive regulation by host of viral transcription(GO:0043923) |
| 0.0 |
0.2 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 |
0.1 |
GO:0048680 |
positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 |
0.0 |
GO:0010652 |
regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 |
0.1 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.0 |
0.4 |
GO:0032012 |
regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.5 |
GO:0003298 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 |
0.4 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 |
0.1 |
GO:0008628 |
hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 |
0.1 |
GO:2000272 |
negative regulation of receptor activity(GO:2000272) |
| 0.0 |
0.1 |
GO:0097017 |
renal protein absorption(GO:0097017) |
| 0.0 |
0.0 |
GO:0007439 |
ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 |
0.0 |
GO:0002731 |
negative regulation of dendritic cell cytokine production(GO:0002731) |
| 0.0 |
0.4 |
GO:0007130 |
synaptonemal complex assembly(GO:0007130) |
| 0.0 |
0.1 |
GO:0048254 |
snoRNA localization(GO:0048254) |
| 0.0 |
0.0 |
GO:0010701 |
positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 |
0.5 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 |
0.3 |
GO:0051601 |
exocyst localization(GO:0051601) |
| 0.0 |
0.6 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
1.3 |
GO:0090502 |
RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 |
0.1 |
GO:0060674 |
placenta blood vessel development(GO:0060674) |
| 0.0 |
0.4 |
GO:0030252 |
growth hormone secretion(GO:0030252) |
| 0.0 |
0.1 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.0 |
0.1 |
GO:0035553 |
oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 |
0.1 |
GO:1902445 |
regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 |
0.2 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 |
0.1 |
GO:0097089 |
methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 |
0.0 |
GO:0021526 |
medial motor column neuron differentiation(GO:0021526) |
| 0.0 |
0.2 |
GO:0002043 |
blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 |
0.7 |
GO:0006198 |
cAMP catabolic process(GO:0006198) |
| 0.0 |
0.0 |
GO:0021823 |
cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 |
0.1 |
GO:1900119 |
positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 |
0.1 |
GO:0033033 |
negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.0 |
0.0 |
GO:0044336 |
canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 |
0.4 |
GO:0045581 |
negative regulation of T cell differentiation(GO:0045581) |
| 0.0 |
0.1 |
GO:0048865 |
stem cell fate commitment(GO:0048865) |
| 0.0 |
0.0 |
GO:1904637 |
response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) negative regulation of synapse maturation(GO:2000297) |
| 0.0 |
0.0 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 |
0.2 |
GO:0090084 |
negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 |
0.1 |
GO:0014028 |
notochord formation(GO:0014028) |
| 0.0 |
0.1 |
GO:0015798 |
myo-inositol transport(GO:0015798) |
| 0.0 |
0.4 |
GO:0009755 |
hormone-mediated signaling pathway(GO:0009755) |
| 0.0 |
0.0 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 |
0.3 |
GO:0030828 |
positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 |
0.2 |
GO:0090385 |
phagosome-lysosome fusion(GO:0090385) |
| 0.0 |
0.2 |
GO:0016446 |
somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 |
0.1 |
GO:1900038 |
negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 |
0.1 |
GO:0071461 |
cellular response to redox state(GO:0071461) |
| 0.0 |
0.1 |
GO:1990502 |
dense core granule maturation(GO:1990502) |
| 0.0 |
0.2 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 |
0.1 |
GO:0000066 |
mitochondrial ornithine transport(GO:0000066) |
| 0.0 |
0.1 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 |
0.1 |
GO:0031580 |
membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 |
0.2 |
GO:0090360 |
platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 |
0.1 |
GO:0090200 |
positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 |
0.1 |
GO:1903025 |
regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 |
0.1 |
GO:0015793 |
glycerol transport(GO:0015793) |
| 0.0 |
0.2 |
GO:0010764 |
negative regulation of fibroblast migration(GO:0010764) |
| 0.0 |
0.1 |
GO:0021590 |
cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 |
0.1 |
GO:0044778 |
meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 |
0.1 |
GO:0021615 |
glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 |
0.1 |
GO:0031666 |
positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 |
0.0 |
GO:0003064 |
regulation of heart rate by hormone(GO:0003064) |
| 0.0 |
0.1 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.0 |
0.1 |
GO:0075525 |
viral translational termination-reinitiation(GO:0075525) |
| 0.0 |
0.1 |
GO:0051708 |
intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 |
0.2 |
GO:1900017 |
positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 |
0.1 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 |
0.2 |
GO:0019371 |
cyclooxygenase pathway(GO:0019371) |
| 0.0 |
0.0 |
GO:1904245 |
modification by virus of host mRNA processing(GO:0046778) regulation of polynucleotide adenylyltransferase activity(GO:1904245) positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 |
0.0 |
GO:0046668 |
regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 |
0.0 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 |
0.2 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) |
| 0.0 |
0.1 |
GO:1903027 |
regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 |
0.2 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.0 |
0.1 |
GO:0031087 |
deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 |
0.1 |
GO:0061470 |
T follicular helper cell differentiation(GO:0061470) |
| 0.0 |
0.3 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 |
0.0 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.0 |
0.2 |
GO:0016576 |
histone dephosphorylation(GO:0016576) |
| 0.0 |
0.0 |
GO:0090214 |
spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 |
0.0 |
GO:1902460 |
transforming growth factor beta activation(GO:0036363) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 |
0.0 |
GO:1900151 |
regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 |
0.4 |
GO:0015718 |
monocarboxylic acid transport(GO:0015718) |
| 0.0 |
0.1 |
GO:0033227 |
dsRNA transport(GO:0033227) |
| 0.0 |
0.3 |
GO:1901685 |
glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 |
0.1 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 |
0.6 |
GO:0015949 |
nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 |
0.2 |
GO:0070498 |
interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 |
0.4 |
GO:0042461 |
photoreceptor cell development(GO:0042461) |
| 0.0 |
0.0 |
GO:0034224 |
cellular response to zinc ion starvation(GO:0034224) |
| 0.0 |
0.2 |
GO:0032793 |
positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 |
0.3 |
GO:0006309 |
apoptotic DNA fragmentation(GO:0006309) |
| 0.0 |
0.1 |
GO:0006689 |
ganglioside catabolic process(GO:0006689) |
| 0.0 |
0.0 |
GO:0032287 |
peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 |
0.2 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 |
0.2 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.4 |
GO:0071800 |
podosome assembly(GO:0071800) |
| 0.0 |
0.1 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 |
0.3 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.0 |
0.2 |
GO:0042340 |
keratan sulfate catabolic process(GO:0042340) |
| 0.0 |
0.0 |
GO:0044027 |
hypermethylation of CpG island(GO:0044027) |
| 0.0 |
0.2 |
GO:0034035 |
purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 |
0.2 |
GO:0051256 |
mitotic spindle midzone assembly(GO:0051256) |
| 0.0 |
0.1 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 |
0.1 |
GO:0036111 |
very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 |
0.2 |
GO:0046500 |
S-adenosylmethionine metabolic process(GO:0046500) |
| 0.0 |
0.1 |
GO:0003406 |
retinal pigment epithelium development(GO:0003406) |