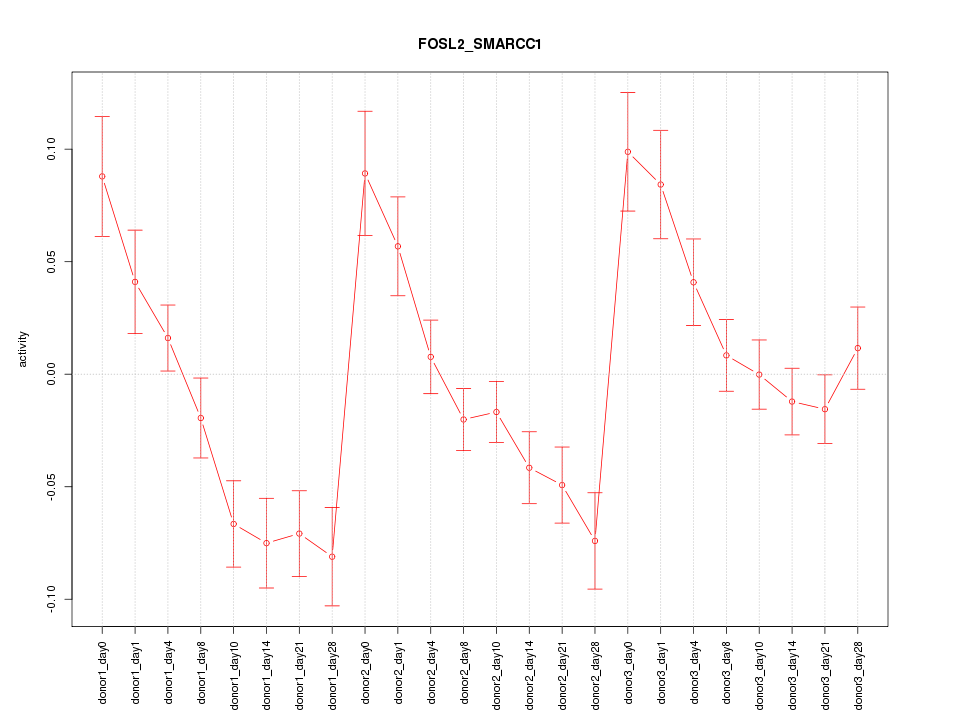
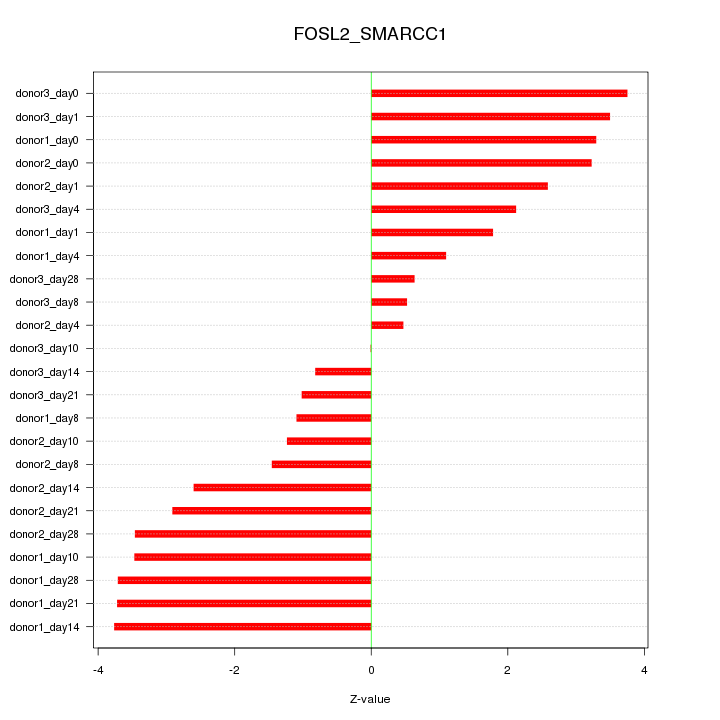
Motif ID: FOSL2_SMARCC1
Z-value: 2.514
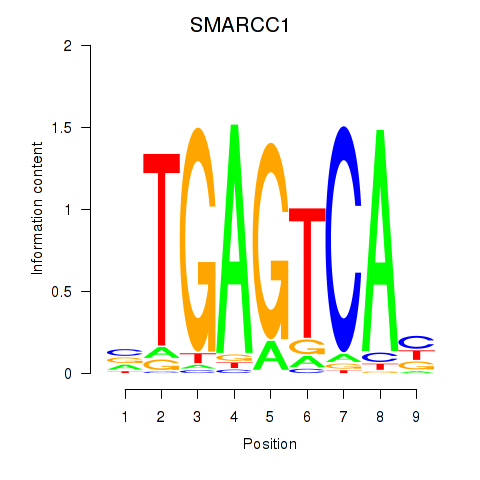
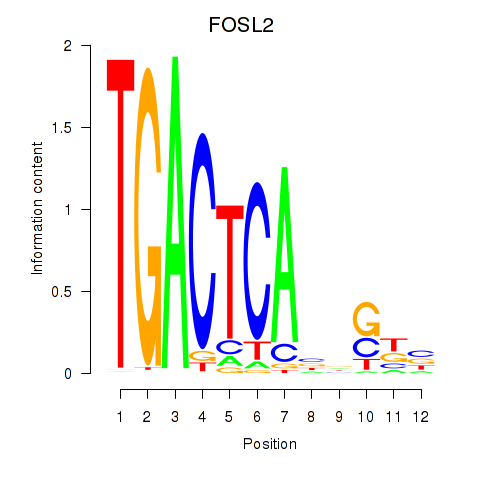
Transcription factors associated with FOSL2_SMARCC1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| FOSL2 | ENSG00000075426.7 | FOSL2 |
| SMARCC1 | ENSG00000173473.6 | SMARCC1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOSL2 | hg19_v2_chr2_+_28618532_28618610 | 0.86 | 9.6e-08 | Click! |
| SMARCC1 | hg19_v2_chr3_-_47823298_47823423 | -0.38 | 6.5e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 13.1 | GO:0033123 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 3.2 | 19.5 | GO:0007296 | vitellogenesis(GO:0007296) |
| 3.2 | 12.7 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 3.1 | 18.4 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 2.8 | 14.0 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 2.8 | 11.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 2.6 | 7.7 | GO:1904298 | positive regulation of neutrophil degranulation(GO:0043315) cellular response to gravity(GO:0071258) positive regulation of neutrophil activation(GO:1902565) regulation of transcytosis(GO:1904298) positive regulation of transcytosis(GO:1904300) regulation of maternal process involved in parturition(GO:1904301) positive regulation of maternal process involved in parturition(GO:1904303) response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904316) cellular response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904317) |
| 2.3 | 66.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 2.2 | 11.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 1.8 | 14.8 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 1.8 | 7.4 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 1.8 | 10.6 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 1.7 | 5.1 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 1.7 | 43.8 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 1.3 | 3.9 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 1.3 | 3.9 | GO:0002384 | hepatic immune response(GO:0002384) |
| 1.2 | 3.6 | GO:0048627 | myoblast development(GO:0048627) |
| 1.1 | 20.3 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 1.1 | 16.9 | GO:0001660 | fever generation(GO:0001660) |
| 1.1 | 3.4 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 1.1 | 7.7 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 1.1 | 5.4 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 1.0 | 1.9 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.9 | 3.8 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.9 | 4.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.8 | 2.5 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.8 | 4.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.8 | 3.3 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.8 | 2.4 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.7 | 3.4 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.7 | 4.6 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.6 | 2.5 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.6 | 3.1 | GO:0060356 | leucine import(GO:0060356) |
| 0.6 | 1.8 | GO:0045658 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.6 | 2.3 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.6 | 2.9 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.6 | 2.9 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.6 | 4.5 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.5 | 2.2 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.5 | 1.6 | GO:0060545 | CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) positive regulation of necroptotic process(GO:0060545) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.5 | 2.6 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.5 | 3.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.5 | 6.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.5 | 5.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.5 | 0.5 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.5 | 7.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.5 | 2.4 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.5 | 1.4 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.5 | 1.4 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.5 | 1.4 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.4 | 3.6 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.4 | 2.7 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.4 | 7.4 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.4 | 2.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.4 | 1.7 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.4 | 2.8 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.4 | 1.2 | GO:0098759 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.4 | 1.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.4 | 1.5 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.4 | 3.8 | GO:0032962 | regulation of inositol trisphosphate biosynthetic process(GO:0032960) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.4 | 10.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.4 | 2.9 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.4 | 5.1 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.4 | 3.2 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.4 | 1.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.4 | 1.4 | GO:1902722 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) positive regulation of prolactin secretion(GO:1902722) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.4 | 1.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.3 | 1.7 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) regulation of determination of dorsal identity(GO:2000015) |
| 0.3 | 3.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.3 | 3.7 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.3 | 1.0 | GO:0060721 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.3 | 1.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.3 | 1.0 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) smooth muscle hyperplasia(GO:0014806) |
| 0.3 | 1.9 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.3 | 1.0 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.3 | 3.2 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.3 | 1.2 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) melanocyte proliferation(GO:0097325) |
| 0.3 | 1.2 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.3 | 0.9 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.3 | 2.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.3 | 0.9 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.3 | 7.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.3 | 0.9 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.3 | 1.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.3 | 4.3 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.3 | 1.7 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.3 | 4.2 | GO:0044351 | macropinocytosis(GO:0044351) phagosome-lysosome fusion(GO:0090385) |
| 0.3 | 3.3 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.3 | 0.8 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.3 | 0.8 | GO:0071451 | cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.3 | 34.9 | GO:0070268 | cornification(GO:0070268) |
| 0.3 | 2.1 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.3 | 0.8 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.3 | 1.0 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.3 | 5.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.3 | 0.3 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.3 | 2.8 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.2 | 3.7 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.2 | 0.7 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.2 | 1.7 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.2 | 2.0 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.2 | 2.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.2 | 12.1 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 9.4 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.2 | 0.7 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.2 | 0.9 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.2 | 3.7 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.2 | 0.7 | GO:0032679 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.2 | 1.1 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.2 | 0.7 | GO:2000910 | negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 0.2 | 1.1 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.2 | 2.5 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.2 | 1.4 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.2 | 1.4 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 0.6 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.2 | 1.9 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.2 | 5.2 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 0.6 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.2 | 2.0 | GO:1905049 | negative regulation of metallopeptidase activity(GO:1905049) |
| 0.2 | 2.9 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.2 | 1.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.2 | 8.8 | GO:0043486 | histone exchange(GO:0043486) |
| 0.2 | 0.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.5 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.2 | 0.4 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.2 | 0.5 | GO:0050894 | determination of affect(GO:0050894) |
| 0.2 | 0.9 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.2 | 0.5 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.2 | 0.7 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.2 | 2.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 1.5 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.2 | 0.3 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.2 | 0.8 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.2 | 10.4 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.2 | 1.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 0.5 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.2 | 15.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.2 | 1.6 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.2 | 2.4 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 3.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.4 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.1 | 1.0 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.9 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.1 | 1.9 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.1 | 2.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 2.2 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.1 | 0.8 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.1 | 0.4 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.1 | 0.7 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 2.0 | GO:2001020 | regulation of response to DNA damage stimulus(GO:2001020) |
| 0.1 | 4.0 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 6.3 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 0.9 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 0.5 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 1.6 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 2.4 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 1.7 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 2.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.5 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 6.5 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 0.5 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 8.2 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) |
| 0.1 | 0.7 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 2.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.2 | GO:0090024 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.6 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.1 | 2.8 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 3.7 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.1 | 5.5 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.1 | 0.6 | GO:0010626 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 | 0.5 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 1.7 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 0.6 | GO:1903401 | L-lysine transmembrane transport(GO:1903401) |
| 0.1 | 1.6 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.4 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 2.9 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 0.5 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 1.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 2.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 1.5 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 1.7 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 2.8 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.9 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 1.0 | GO:0035588 | G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 0.7 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 0.5 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.1 | 4.6 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 0.6 | GO:0010919 | regulation of inositol phosphate biosynthetic process(GO:0010919) positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 0.8 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.1 | GO:0014040 | positive regulation of Schwann cell differentiation(GO:0014040) |
| 0.1 | 1.9 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 2.1 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.1 | 1.2 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 0.5 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 3.6 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 0.4 | GO:0060737 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.1 | 0.2 | GO:2000861 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.1 | 0.2 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.1 | 0.9 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.1 | 1.6 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.2 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.1 | 1.3 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.1 | 0.5 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.1 | 0.2 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.1 | 1.1 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.1 | 0.3 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 1.2 | GO:0050777 | negative regulation of immune response(GO:0050777) |
| 0.1 | 1.1 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.1 | 4.2 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.1 | 0.7 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 1.2 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.4 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.3 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) negative regulation of mitophagy(GO:1903147) |
| 0.0 | 3.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 1.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 5.8 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 1.9 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 2.6 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 | 0.5 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 1.0 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.4 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.9 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:1903597 | regulation of gap junction assembly(GO:1903596) negative regulation of gap junction assembly(GO:1903597) positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.0 | 1.2 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 1.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.1 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.5 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 1.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 1.3 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 1.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 1.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 2.2 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.4 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.1 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.2 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.5 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.0 | 0.8 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 2.3 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.0 | 0.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 1.4 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.5 | GO:0035729 | cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 0.1 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.9 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.6 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 3.0 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.0 | 1.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.0 | 0.2 | GO:0071651 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 1.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.6 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.6 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.4 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.8 | GO:0006520 | cellular amino acid metabolic process(GO:0006520) |
| 0.0 | 1.0 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.1 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 1.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.3 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.3 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.0 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.0 | 1.1 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 0.0 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 1.2 | GO:0006901 | vesicle coating(GO:0006901) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 1.9 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.8 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.8 | GO:0003231 | cardiac ventricle development(GO:0003231) |
| 0.0 | 1.9 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 1.7 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 2.5 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.2 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 1.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.4 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.5 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 1.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.0 | GO:0010756 | regulation of plasminogen activation(GO:0010755) positive regulation of plasminogen activation(GO:0010756) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.6 | 52.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 2.9 | 8.8 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 2.4 | 43.9 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 1.9 | 7.6 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 1.4 | 10.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 1.4 | 11.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 1.1 | 5.4 | GO:0032449 | CBM complex(GO:0032449) |
| 1.0 | 5.9 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 1.0 | 3.9 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.8 | 8.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.8 | 3.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.7 | 2.2 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.7 | 4.6 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.6 | 22.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.6 | 1.7 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.6 | 36.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.5 | 3.2 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.5 | 4.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.5 | 1.8 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.4 | 4.4 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.4 | 8.8 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.4 | 8.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.4 | 1.4 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.3 | 4.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.3 | 1.4 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.3 | 1.6 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.3 | 4.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 0.9 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.2 | 9.5 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 4.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.2 | 2.9 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 28.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 2.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 2.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 19.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 2.4 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.2 | 0.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.2 | 2.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 0.6 | GO:0036338 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.2 | 18.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 8.0 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 0.9 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 0.6 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 1.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 2.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.8 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 1.5 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 32.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 2.9 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 1.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.5 | GO:0002081 | inner acrosomal membrane(GO:0002079) outer acrosomal membrane(GO:0002081) |
| 0.1 | 2.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.9 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.5 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 3.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 3.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 1.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 2.3 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 10.5 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 3.7 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 3.3 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.7 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.1 | 0.7 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 2.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.8 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 0.5 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 2.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.9 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 4.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.0 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 0.8 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.3 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.1 | 4.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 17.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.1 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 1.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 4.6 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.5 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.9 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 3.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 4.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.3 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 1.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 9.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 4.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 1.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.5 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 3.3 | GO:0031253 | cell projection membrane(GO:0031253) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 12.2 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 49.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.6 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.3 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 22.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.3 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.0 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 4.0 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.8 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.5 | GO:0043209 | myelin sheath(GO:0043209) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 43.9 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 4.0 | 12.0 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 2.5 | 12.7 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 2.5 | 7.4 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 2.2 | 11.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 1.5 | 7.4 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 1.4 | 4.3 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 1.4 | 10.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 1.4 | 4.1 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 1.3 | 5.0 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 1.1 | 13.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 1.0 | 10.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 1.0 | 6.8 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.9 | 2.6 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.8 | 5.9 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.7 | 2.8 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.7 | 3.9 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.6 | 11.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.6 | 21.9 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.5 | 2.2 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.5 | 2.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.5 | 1.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.5 | 9.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.5 | 2.4 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.5 | 1.4 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.5 | 1.4 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.4 | 1.2 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.4 | 1.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.4 | 3.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.4 | 1.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.4 | 1.4 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.4 | 7.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.3 | 3.8 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.3 | 1.7 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.3 | 3.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 2.3 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.3 | 4.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.3 | 4.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 5.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.3 | 1.0 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.3 | 3.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.3 | 1.9 | GO:0042806 | fucose binding(GO:0042806) |
| 0.3 | 11.8 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.3 | 1.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 0.9 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.3 | 2.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.3 | 5.4 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.3 | 1.7 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.3 | 1.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.3 | 7.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.3 | 1.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.3 | 1.6 | GO:0042835 | BRE binding(GO:0042835) |
| 0.3 | 0.8 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.3 | 4.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 1.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.2 | 1.0 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.2 | 2.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 1.4 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 0.9 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.2 | 1.9 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 1.6 | GO:0050816 | phosphoserine binding(GO:0050815) phosphothreonine binding(GO:0050816) |
| 0.2 | 0.8 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 3.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 0.8 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.2 | 3.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 13.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 2.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 4.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 2.0 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.2 | 2.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 0.5 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.2 | 0.5 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.2 | 55.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 5.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 0.7 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.2 | 0.5 | GO:0031711 | angiotensin type I receptor activity(GO:0001596) bradykinin receptor binding(GO:0031711) |
| 0.2 | 20.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 3.6 | GO:0016918 | retinal binding(GO:0016918) |
| 0.2 | 0.5 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.2 | 0.8 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.2 | 3.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.4 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 6.6 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 4.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.4 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.1 | 1.9 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 4.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.5 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.1 | 1.8 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 1.4 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.9 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 1.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 8.8 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 0.6 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 2.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.6 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 20.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 3.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 1.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 2.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 3.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 1.8 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 77.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 0.7 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 2.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 14.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 5.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.7 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.6 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 1.7 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 2.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 2.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.2 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.1 | 1.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 2.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 5.2 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.1 | 0.7 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 4.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 0.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 2.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.8 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.2 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 1.6 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.1 | 3.4 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.6 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 1.0 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.3 | GO:0008200 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.2 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.1 | 0.2 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.1 | 1.0 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 1.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.2 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.8 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 0.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.8 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.4 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 4.3 | GO:0008238 | exopeptidase activity(GO:0008238) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.2 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.6 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 1.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 1.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 2.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 2.5 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.0 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 3.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 4.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.9 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.5 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.5 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 2.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 2.4 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.3 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 1.8 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 1.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.7 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 1.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.3 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 5.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 1.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 68.0 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.6 | 41.8 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.5 | 15.4 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.5 | 14.9 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.4 | 11.2 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.3 | 15.1 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.3 | 21.8 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 9.8 | PID_IL1_PATHWAY | IL1-mediated signaling events |
| 0.2 | 5.1 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 4.6 | PID_S1P_S1P1_PATHWAY | S1P1 pathway |
| 0.2 | 46.8 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 9.4 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.2 | 3.5 | PID_ECADHERIN_KERATINOCYTE_PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.7 | SA_TRKA_RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 7.6 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 8.9 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 5.3 | PID_CD8_TCR_DOWNSTREAM_PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 2.7 | SA_FAS_SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 25.7 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 7.1 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.5 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 4.1 | PID_BCR_5PATHWAY | BCR signaling pathway |
| 0.1 | 1.9 | PID_AMB2_NEUTROPHILS_PATHWAY | amb2 Integrin signaling |
| 0.1 | 5.6 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.1 | 0.7 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.1 | 1.9 | PID_ECADHERIN_NASCENT_AJ_PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 21.3 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 2.3 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.2 | PID_GMCSF_PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 1.4 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.7 | PID_PI3K_PLC_TRK_PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.4 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 2.1 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.4 | PID_PI3KCI_PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.6 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.7 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.0 | 2.1 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | PID_CXCR4_PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 2.1 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.4 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.6 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | PID_IL8_CXCR1_PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.3 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.4 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.4 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | PID_THROMBIN_PAR4_PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.5 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID_CERAMIDE_PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.5 | PID_P75_NTR_PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.8 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 12.7 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.5 | 14.2 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.5 | 59.1 | REACTOME_CELL_JUNCTION_ORGANIZATION | Genes involved in Cell junction organization |
| 0.4 | 10.1 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.3 | 6.5 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 10.3 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.3 | 22.3 | REACTOME_INTEGRIN_ALPHAIIB_BETA3_SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.3 | 4.5 | REACTOME_REVERSIBLE_HYDRATION_OF_CARBON_DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 15.9 | REACTOME_IL1_SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.3 | 13.0 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 0.4 | REACTOME_P75NTR_RECRUITS_SIGNALLING_COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.2 | 5.1 | REACTOME_REGULATION_OF_IFNA_SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 3.9 | REACTOME_IL_6_SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 7.4 | REACTOME_POST_TRANSLATIONAL_MODIFICATION_SYNTHESIS_OF_GPI_ANCHORED_PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.2 | 3.0 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELL_ADHESION_PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 3.4 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 4.6 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 2.7 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 2.7 | REACTOME_AKT_PHOSPHORYLATES_TARGETS_IN_THE_CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 1.9 | REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 4.6 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 4.9 | REACTOME_ACTIVATION_OF_GENES_BY_ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 2.2 | REACTOME_SIGNALING_BY_CONSTITUTIVELY_ACTIVE_EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 3.2 | REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 5.0 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 3.6 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 3.0 | REACTOME_RIP_MEDIATED_NFKB_ACTIVATION_VIA_DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 7.4 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.1 | 3.9 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.0 | REACTOME_ACTIVATED_POINT_MUTANTS_OF_FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 5.6 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.0 | REACTOME_TETRAHYDROBIOPTERIN_BH4_SYNTHESIS_RECYCLING_SALVAGE_AND_REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.1 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.6 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.9 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.4 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.1 | 1.7 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.1 | 2.5 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.1 | 2.0 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.0 | REACTOME_RECYCLING_OF_BILE_ACIDS_AND_SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.2 | REACTOME_G_ALPHA_Z_SIGNALLING_EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 3.8 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 0.8 | REACTOME_ER_PHAGOSOME_PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 1.7 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 4.8 | REACTOME_CELL_SURFACE_INTERACTIONS_AT_THE_VASCULAR_WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 1.7 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.4 | REACTOME_PROTEOLYTIC_CLEAVAGE_OF_SNARE_COMPLEX_PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.0 | REACTOME_ADENYLATE_CYCLASE_INHIBITORY_PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 3.2 | REACTOME_GLUCOSE_METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 1.9 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 11.7 | REACTOME_G_ALPHA_Q_SIGNALLING_EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 2.6 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.0 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.9 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.8 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.7 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.8 | REACTOME_TRAFFICKING_OF_AMPA_RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.6 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.1 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.2 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 3.3 | REACTOME_PPARA_ACTIVATES_GENE_EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 5.2 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME_PREFOLDIN_MEDIATED_TRANSFER_OF_SUBSTRATE_TO_CCT_TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.5 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.7 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.1 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.9 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.5 | REACTOME_OLFACTORY_SIGNALING_PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.9 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.7 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.2 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.9 | REACTOME_NITRIC_OXIDE_STIMULATES_GUANYLATE_CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.3 | REACTOME_SMAD2_SMAD3_SMAD4_HETEROTRIMER_REGULATES_TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.7 | REACTOME_PHASE_II_CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.0 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.0 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.6 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.0 | 0.4 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME_ORGANIC_CATION_ANION_ZWITTERION_TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.1 | REACTOME_LIPOPROTEIN_METABOLISM | Genes involved in Lipoprotein metabolism |