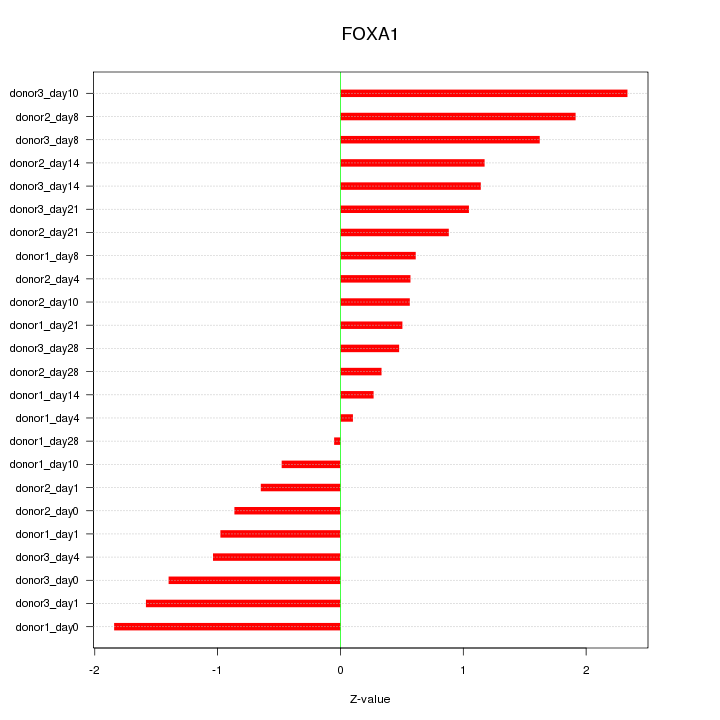
Motif ID: FOXA1
Z-value: 1.105
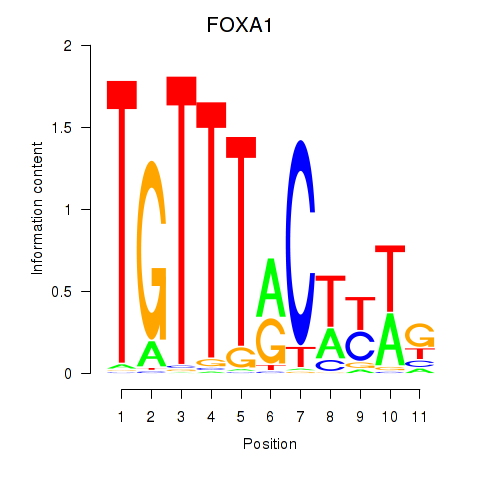
Transcription factors associated with FOXA1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| FOXA1 | ENSG00000129514.4 | FOXA1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXA1 | hg19_v2_chr14_-_38064198_38064239 | 0.41 | 4.8e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.5 | GO:0042710 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 1.5 | 4.4 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 1.4 | 4.2 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.8 | 3.4 | GO:0072134 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme morphogenesis(GO:0072134) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) allantois development(GO:1905069) |
| 0.6 | 1.7 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.4 | 1.8 | GO:0018032 | protein amidation(GO:0018032) |
| 0.3 | 3.0 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.3 | 0.9 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.3 | 1.4 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.3 | 1.9 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.3 | 1.6 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.2 | 1.2 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.2 | 0.8 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.2 | 4.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 0.6 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.2 | 3.4 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.2 | 0.7 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.2 | 0.5 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.2 | 0.8 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.2 | 2.5 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.2 | 2.6 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 2.1 | GO:0021527 | ventral spinal cord interneuron differentiation(GO:0021514) spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 1.0 | GO:2000334 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 11.7 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 3.4 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 1.0 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 4.9 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 0.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.5 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.3 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 | 0.5 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 | 0.5 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 2.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.4 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 1.0 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.4 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.8 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.2 | GO:1901297 | blood vessel lumenization(GO:0072554) positive regulation of ephrin receptor signaling pathway(GO:1901189) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.8 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.1 | 0.3 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.5 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.6 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.4 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) |
| 0.0 | 0.6 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 1.0 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 1.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.9 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.4 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.3 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.2 | GO:1904479 | regulation of isoprenoid metabolic process(GO:0019747) negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:1903756 | positive regulation of dendritic spine maintenance(GO:1902952) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.3 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.6 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.9 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.9 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.6 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.2 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.9 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 1.9 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 0.2 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 1.9 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 1.6 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.4 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.2 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 1.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.0 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 1.5 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.7 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.3 | 0.8 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.2 | 0.6 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.9 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.3 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.2 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.1 | 0.2 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.5 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.3 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 2.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 1.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.9 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.3 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 2.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 3.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 2.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 1.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 12.9 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.6 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0016160 | amylase activity(GO:0016160) |
| 0.4 | 1.8 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.4 | 1.7 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.4 | 1.2 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.4 | 4.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.3 | 2.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 0.8 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.3 | 3.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 0.9 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.2 | 0.6 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.2 | 1.4 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.2 | 0.7 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 1.0 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.7 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.5 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.1 | 0.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.2 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.5 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 0.2 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.9 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 1.7 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 10.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.2 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.2 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.1 | 2.1 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.6 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.6 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 5.1 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.1 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 5.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 1.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.6 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 0.1 | 4.2 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 4.2 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 3.0 | PID_ECADHERIN_NASCENT_AJ_PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 4.9 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.6 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.0 | 3.9 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | PID_CD40_PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.3 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.2 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 2.7 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.9 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.4 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | REACTOME_DIGESTION_OF_DIETARY_CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 5.4 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.7 | REACTOME_ETHANOL_OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.5 | REACTOME_IRON_UPTAKE_AND_TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 1.6 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.0 | REACTOME_DSCAM_INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.6 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.5 | REACTOME_CREATION_OF_C4_AND_C2_ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.8 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.7 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.9 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.2 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.8 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.2 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.2 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME_RECYCLING_OF_BILE_ACIDS_AND_SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 4.5 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.8 | REACTOME_COMPLEMENT_CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.5 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.3 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.9 | REACTOME_GLUCOSE_TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.2 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME_IRAK1_RECRUITS_IKK_COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.6 | REACTOME_SIGNALING_BY_NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 0.3 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |