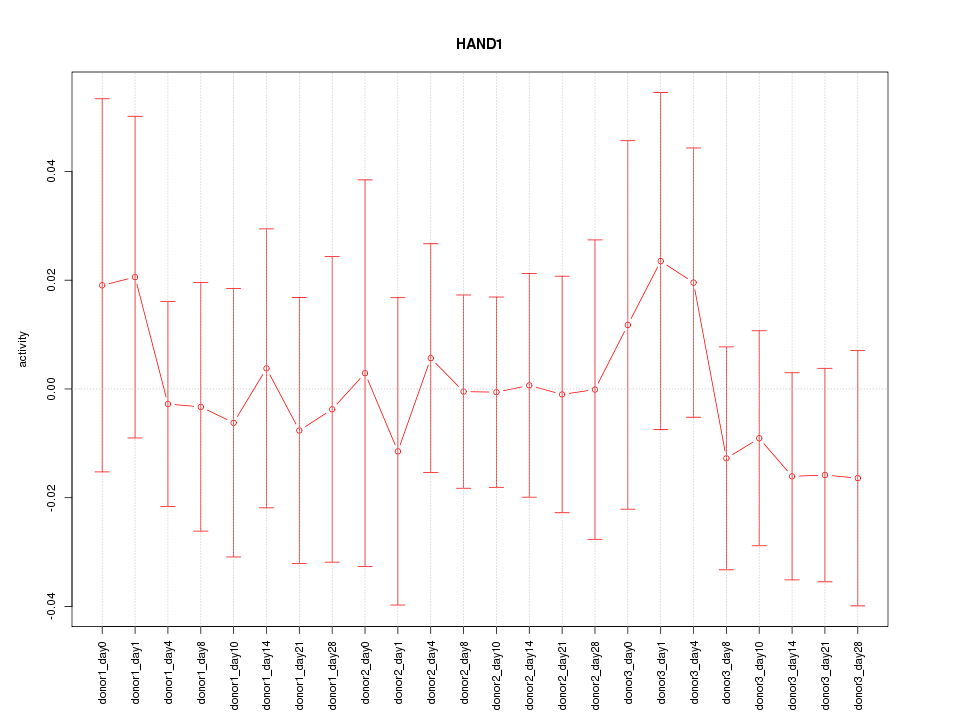
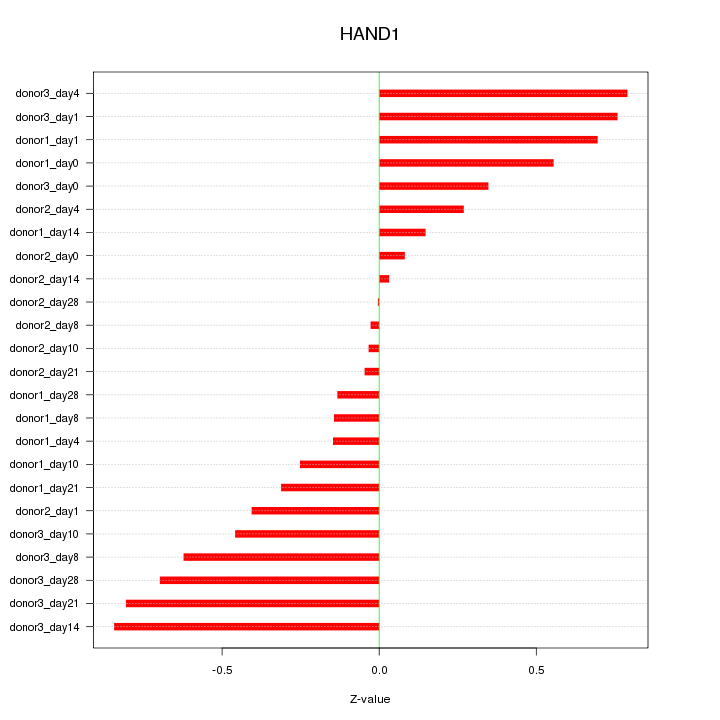
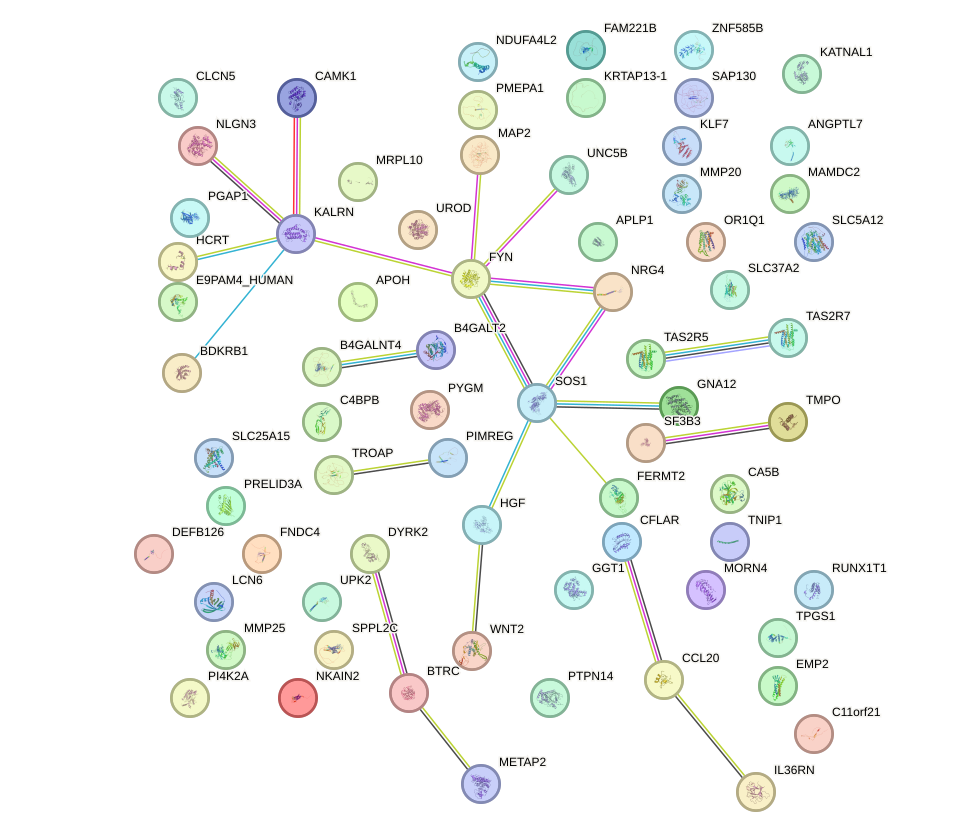
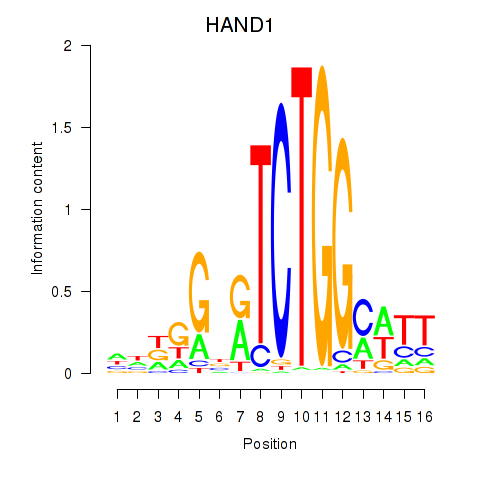
Motif ID: HAND1
Z-value: 0.459
Transcription factors associated with HAND1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| HAND1 | ENSG00000113196.2 | HAND1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HAND1 | hg19_v2_chr5_-_153857819_153857824 | -0.09 | 6.9e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 1.4 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.4 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 0.5 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 0.2 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.1 | 0.5 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.1 | 0.9 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.2 | GO:2000338 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) positive regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000340) |
| 0.1 | 0.6 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.5 | GO:1903027 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.5 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.3 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.4 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.1 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.0 | 0.1 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.1 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.1 | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.8 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.0 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.0 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.3 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.6 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 1.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.3 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.1 | 0.6 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.1 | 0.8 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.2 | 0.6 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.2 | 1.4 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.5 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 0.4 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.2 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 0.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.2 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 1.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA_FAS_SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.4 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.4 | REACTOME_PLATELET_ADHESION_TO_EXPOSED_COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.9 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.5 | REACTOME_REGULATION_OF_COMPLEMENT_CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.5 | REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME_CTLA4_INHIBITORY_SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.6 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME_PLATELET_SENSITIZATION_BY_LDL | Genes involved in Platelet sensitization by LDL |