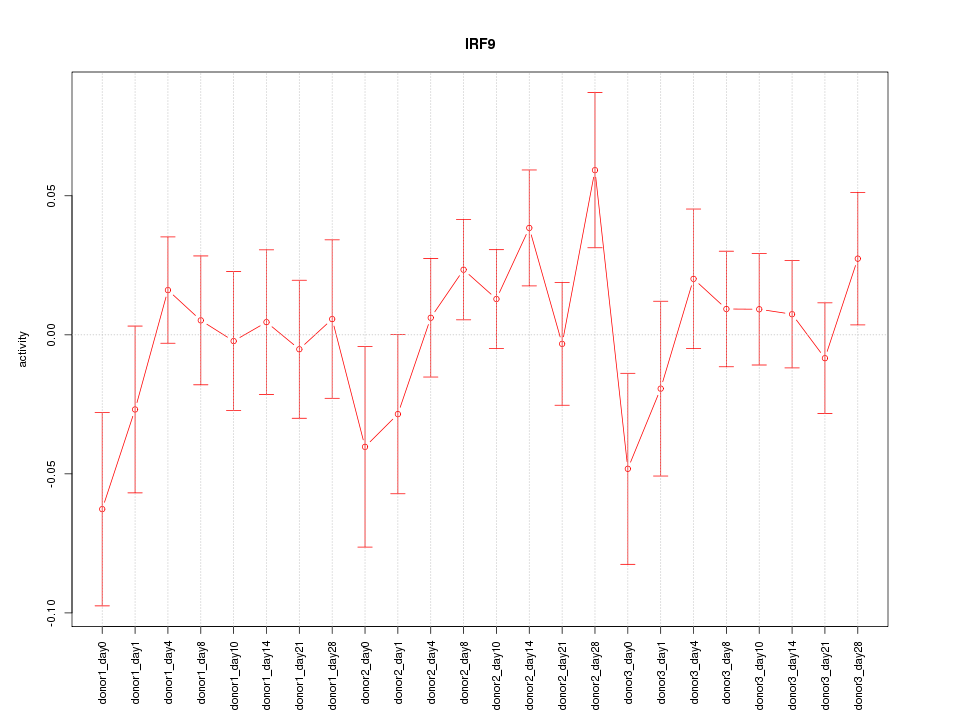
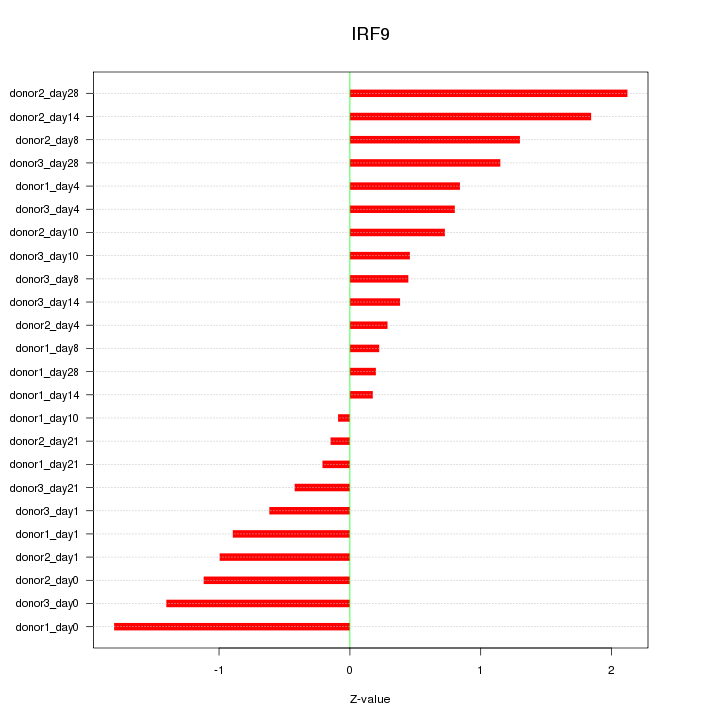
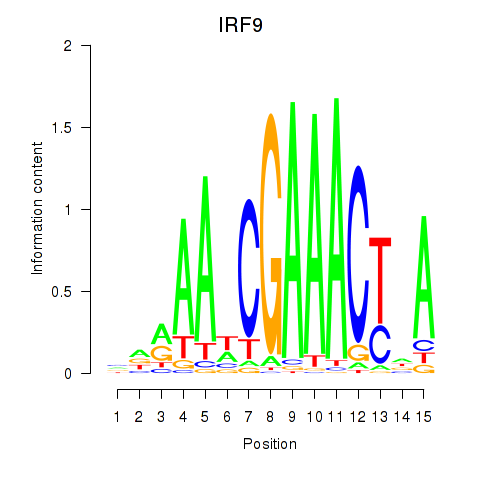
Motif ID: IRF9
Z-value: 0.967
Transcription factors associated with IRF9:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| IRF9 | ENSG00000213928.4 | IRF9 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRF9 | hg19_v2_chr14_+_24630465_24630531 | 0.09 | 6.7e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.8 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 1.2 | 3.6 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.8 | 9.3 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.8 | 2.3 | GO:1901253 | negative regulation of dendritic cell cytokine production(GO:0002731) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.7 | 3.5 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.7 | 2.7 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.6 | 3.1 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.6 | 2.3 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.4 | 1.2 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.3 | 3.0 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.3 | 2.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.3 | 2.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.2 | 0.7 | GO:0050823 | peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.2 | 0.6 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.2 | 0.5 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.2 | 1.5 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.7 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.1 | 1.9 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 0.5 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.1 | 0.9 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.6 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.1 | 3.5 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.1 | 2.6 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.8 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 1.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.5 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.8 | GO:2000111 | senescence-associated heterochromatin focus assembly(GO:0035986) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.1 | 0.5 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.3 | GO:0019075 | virus maturation(GO:0019075) |
| 0.1 | 5.0 | GO:0034340 | response to type I interferon(GO:0034340) |
| 0.0 | 0.4 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.7 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.5 | GO:0043950 | regulation of T cell chemotaxis(GO:0010819) positive regulation of cAMP-mediated signaling(GO:0043950) regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.3 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.2 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 1.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.5 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.3 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.9 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.4 | 2.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 1.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 1.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.8 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 5.8 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 1.0 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 5.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 5.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.7 | GO:0071556 | integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.6 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.8 | 2.3 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.5 | 3.0 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.4 | 3.5 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.3 | 1.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.3 | 2.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 1.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 1.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 1.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 2.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.7 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.1 | 0.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 1.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 2.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 2.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 9.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 0.9 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.5 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 2.7 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.1 | 0.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 1.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 9.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST_TYPE_I_INTERFERON_PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 4.0 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.8 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.8 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID_ALK2_PATHWAY | ALK2 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.5 | REACTOME_ENDOSOMAL_VACUOLAR_PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.3 | 5.0 | REACTOME_NFKB_ACTIVATION_THROUGH_FADD_RIP1_PATHWAY_MEDIATED_BY_CASPASE_8_AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.2 | 20.3 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 3.1 | REACTOME_PURINE_CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 3.2 | REACTOME_TRYPTOPHAN_CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 2.4 | REACTOME_RIG_I_MDA5_MEDIATED_INDUCTION_OF_IFN_ALPHA_BETA_PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 0.8 | REACTOME_SHC1_EVENTS_IN_ERBB4_SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.4 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME_G1_PHASE | Genes involved in G1 Phase |