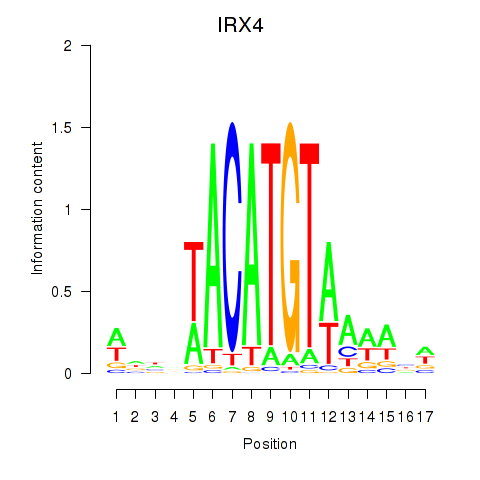
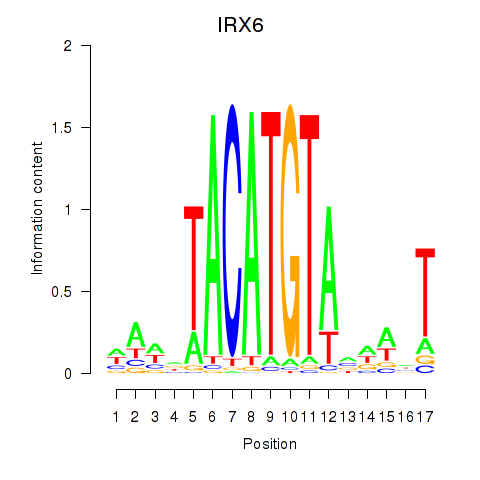
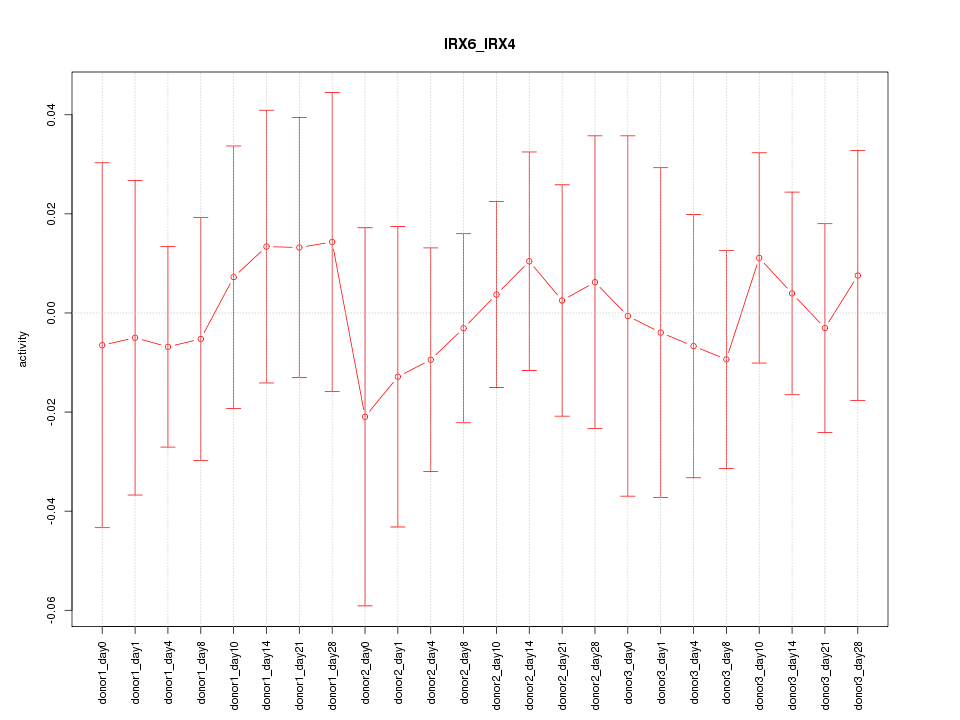
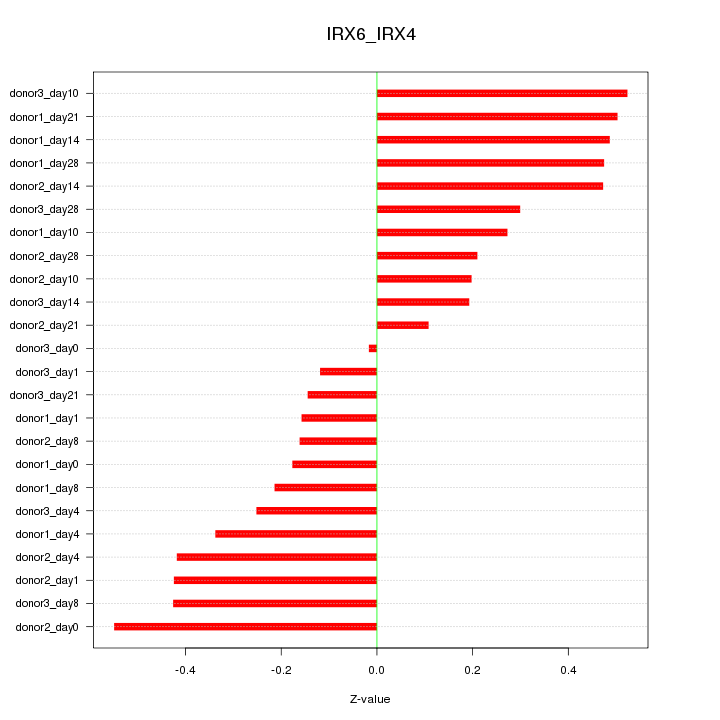
|
chr16_+_777118
|
0.818
|
ENST00000562141.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like
|
|
chr3_-_9994021
|
0.632
|
ENST00000411976.2
ENST00000412055.1
|
PRRT3
|
proline-rich transmembrane protein 3
|
|
chr1_+_63989004
|
0.621
|
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7
|
|
chr21_-_34185944
|
0.516
|
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62
|
|
chr11_+_6897856
|
0.505
|
ENST00000379829.2
|
OR10A4
|
olfactory receptor, family 10, subfamily A, member 4
|
|
chr16_+_777246
|
0.446
|
ENST00000561546.1
ENST00000564545.1
ENST00000389703.3
ENST00000567414.1
ENST00000568141.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like
|
|
chr16_+_776936
|
0.409
|
ENST00000549114.1
ENST00000341413.4
ENST00000562187.1
ENST00000564537.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like
|
|
chr20_+_11008408
|
0.397
|
ENST00000378252.1
|
C20orf187
|
chromosome 20 open reading frame 187
|
|
chr12_+_32655110
|
0.371
|
ENST00000546442.1
ENST00000583694.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr8_+_67976593
|
0.350
|
ENST00000262210.5
ENST00000412460.1
|
CSPP1
|
centrosome and spindle pole associated protein 1
|
|
chr12_-_10282836
|
0.325
|
ENST00000304084.8
ENST00000353231.5
ENST00000525605.1
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chr4_+_41361616
|
0.296
|
ENST00000513024.1
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr6_-_52705641
|
0.270
|
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5
|
|
chr7_-_16844611
|
0.259
|
ENST00000401412.1
ENST00000419304.2
|
AGR2
|
anterior gradient 2
|
|
chr1_+_170904612
|
0.257
|
ENST00000367759.4
ENST00000367758.3
|
MROH9
|
maestro heat-like repeat family member 9
|
|
chr12_-_10282681
|
0.225
|
ENST00000533022.1
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chr3_+_171561127
|
0.223
|
ENST00000334567.5
ENST00000450693.1
|
TMEM212
|
transmembrane protein 212
|
|
chr12_-_10282742
|
0.215
|
ENST00000298523.5
ENST00000396484.2
ENST00000310002.4
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chrX_+_36254051
|
0.204
|
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30
|
|
chr13_-_41768654
|
0.176
|
ENST00000379483.3
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7
|
|
chr17_-_42327236
|
0.167
|
ENST00000399246.2
|
AC003102.1
|
AC003102.1
|
|
chr8_-_17555164
|
0.166
|
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr21_-_34185989
|
0.163
|
ENST00000487113.1
ENST00000382373.4
|
C21orf62
|
chromosome 21 open reading frame 62
|
|
chr11_-_35547572
|
0.152
|
ENST00000378880.2
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr2_+_113829895
|
0.133
|
ENST00000393197.2
|
IL1F10
|
interleukin 1 family, member 10 (theta)
|
|
chr20_+_43990576
|
0.131
|
ENST00000372727.1
ENST00000414310.1
|
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)
|
|
chr19_-_54974894
|
0.130
|
ENST00000333834.4
|
LENG9
|
leukocyte receptor cluster (LRC) member 9
|
|
chr10_+_118350522
|
0.130
|
ENST00000530319.1
ENST00000527980.1
ENST00000471549.1
ENST00000534537.1
|
PNLIPRP1
|
pancreatic lipase-related protein 1
|
|
chr17_-_29641084
|
0.121
|
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr10_+_118350468
|
0.121
|
ENST00000358834.4
ENST00000528052.1
ENST00000442761.1
|
PNLIPRP1
|
pancreatic lipase-related protein 1
|
|
chr2_+_113670548
|
0.119
|
ENST00000263326.3
ENST00000352179.3
ENST00000349806.3
ENST00000353225.3
|
IL37
|
interleukin 37
|
|
chr10_+_90660832
|
0.115
|
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1
|
|
chr6_+_116691001
|
0.109
|
ENST00000537543.1
|
DSE
|
dermatan sulfate epimerase
|
|
chr6_+_140175987
|
0.108
|
ENST00000414038.1
ENST00000431609.1
|
RP5-899B16.1
|
RP5-899B16.1
|
|
chr7_-_92855762
|
0.104
|
ENST00000453812.2
ENST00000394468.2
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr18_-_47376197
|
0.101
|
ENST00000592688.1
|
MYO5B
|
myosin VB
|
|
chr20_+_10199468
|
0.101
|
ENST00000254976.2
ENST00000304886.2
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr17_-_47723943
|
0.096
|
ENST00000510476.1
ENST00000503676.1
|
SPOP
|
speckle-type POZ protein
|
|
chr15_+_41057818
|
0.094
|
ENST00000558467.1
|
GCHFR
|
GTP cyclohydrolase I feedback regulator
|
|
chr2_+_133874577
|
0.090
|
ENST00000596384.1
|
AC011755.1
|
HCG2006742; Protein LOC100996685
|
|
chr15_+_66585879
|
0.088
|
ENST00000319212.4
|
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like
|
|
chr15_+_66585555
|
0.082
|
ENST00000319194.5
ENST00000525134.2
ENST00000441424.2
|
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like
|
|
chr9_-_37465396
|
0.082
|
ENST00000307750.4
|
ZBTB5
|
zinc finger and BTB domain containing 5
|
|
chr16_+_21689835
|
0.076
|
ENST00000286149.4
ENST00000388958.3
|
OTOA
|
otoancorin
|
|
chr4_+_130017268
|
0.074
|
ENST00000425929.1
ENST00000508673.1
ENST00000508622.1
|
C4orf33
|
chromosome 4 open reading frame 33
|
|
chr3_+_53528659
|
0.068
|
ENST00000350061.5
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit
|
|
chr9_-_27573651
|
0.067
|
ENST00000379995.1
ENST00000379997.3
|
C9orf72
|
chromosome 9 open reading frame 72
|
|
chr19_+_50691437
|
0.061
|
ENST00000598205.1
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chr2_-_231989808
|
0.061
|
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled
|
|
chr14_+_56127989
|
0.055
|
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor)
|
|
chr7_-_80141328
|
0.053
|
ENST00000398291.3
|
GNAT3
|
guanine nucleotide binding protein, alpha transducing 3
|
|
chr13_-_46716969
|
0.049
|
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr21_-_34186006
|
0.048
|
ENST00000490358.1
|
C21orf62
|
chromosome 21 open reading frame 62
|
|
chrX_-_15333736
|
0.046
|
ENST00000380470.3
|
ASB11
|
ankyrin repeat and SOCS box containing 11
|
|
chr8_+_84824920
|
0.042
|
ENST00000523678.1
|
RP11-120I21.2
|
RP11-120I21.2
|
|
chr4_+_71458012
|
0.039
|
ENST00000449493.2
|
AMBN
|
ameloblastin (enamel matrix protein)
|
|
chr1_+_53308398
|
0.034
|
ENST00000371528.1
|
ZYG11A
|
zyg-11 family member A, cell cycle regulator
|
|
chr5_+_59783540
|
0.030
|
ENST00000515734.2
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr2_+_58655461
|
0.028
|
ENST00000429095.1
ENST00000429664.1
ENST00000452840.1
|
AC007092.1
|
long intergenic non-protein coding RNA 1122
|
|
chr4_+_113558612
|
0.026
|
ENST00000505034.1
ENST00000324052.6
|
LARP7
|
La ribonucleoprotein domain family, member 7
|
|
chrX_+_15767971
|
0.025
|
ENST00000479740.1
ENST00000454127.2
|
CA5B
|
carbonic anhydrase VB, mitochondrial
|
|
chr1_-_3566590
|
0.024
|
ENST00000424367.1
ENST00000378322.3
|
WRAP73
|
WD repeat containing, antisense to TP73
|
|
chr4_+_71457970
|
0.022
|
ENST00000322937.6
|
AMBN
|
ameloblastin (enamel matrix protein)
|
|
chr12_+_5541267
|
0.020
|
ENST00000423158.3
|
NTF3
|
neurotrophin 3
|
|
chr5_+_175490540
|
0.019
|
ENST00000515817.1
|
FAM153B
|
family with sequence similarity 153, member B
|
|
chr3_-_168865522
|
0.015
|
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr1_-_3566627
|
0.013
|
ENST00000419924.2
ENST00000270708.7
|
WRAP73
|
WD repeat containing, antisense to TP73
|
|
chr8_-_67976509
|
0.007
|
ENST00000518747.1
|
COPS5
|
COP9 signalosome subunit 5
|
|
chr12_-_88423164
|
0.007
|
ENST00000298699.2
ENST00000550553.1
|
C12orf50
|
chromosome 12 open reading frame 50
|
|
chr10_-_115904361
|
0.001
|
ENST00000428953.1
ENST00000543782.1
|
C10orf118
|
chromosome 10 open reading frame 118
|
|
chr6_-_49834240
|
0.001
|
ENST00000335847.4
|
CRISP1
|
cysteine-rich secretory protein 1
|