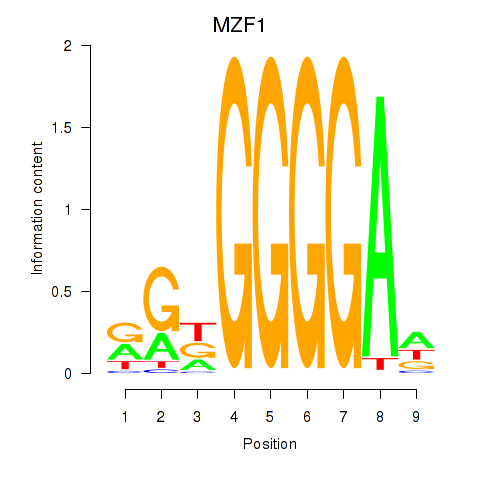
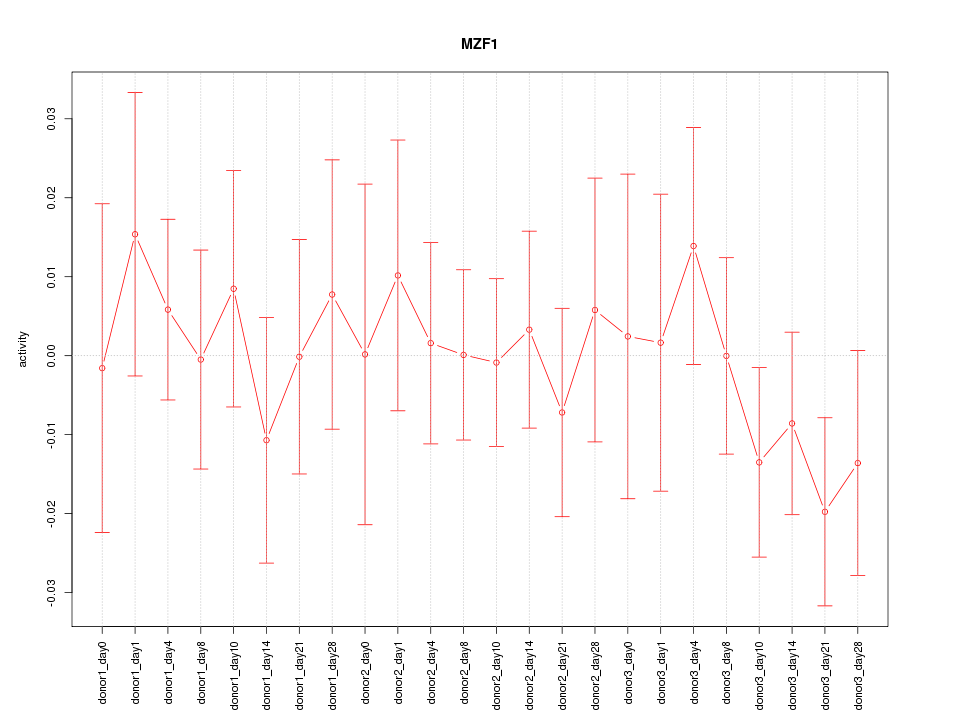
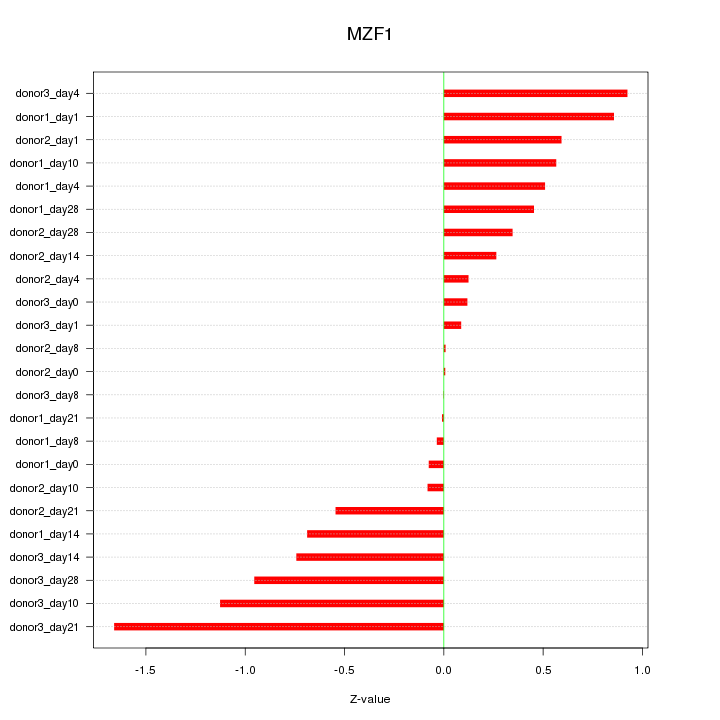
| 0.3 |
1.0 |
GO:0046586 |
regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.2 |
0.5 |
GO:0000412 |
histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.2 |
0.5 |
GO:2000854 |
positive regulation of corticosterone secretion(GO:2000854) |
| 0.2 |
0.6 |
GO:0033123 |
positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.1 |
0.4 |
GO:2001037 |
tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.1 |
0.8 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 |
0.4 |
GO:0052047 |
interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.1 |
0.4 |
GO:1902595 |
regulation of DNA replication origin binding(GO:1902595) |
| 0.1 |
0.6 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 |
0.3 |
GO:0090341 |
negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.1 |
0.6 |
GO:1902617 |
response to fluoride(GO:1902617) |
| 0.1 |
0.3 |
GO:1903860 |
negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.1 |
0.3 |
GO:0018894 |
dibenzo-p-dioxin metabolic process(GO:0018894) cellular alkene metabolic process(GO:0043449) |
| 0.1 |
0.5 |
GO:0061143 |
alveolar primary septum development(GO:0061143) |
| 0.1 |
0.4 |
GO:1903691 |
positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 |
0.4 |
GO:0031117 |
positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 |
0.3 |
GO:0014016 |
neuroblast differentiation(GO:0014016) |
| 0.1 |
0.5 |
GO:0001661 |
conditioned taste aversion(GO:0001661) |
| 0.1 |
0.2 |
GO:0006535 |
cysteine biosynthetic process from serine(GO:0006535) |
| 0.1 |
0.7 |
GO:2000253 |
positive regulation of feeding behavior(GO:2000253) |
| 0.1 |
0.2 |
GO:0032911 |
negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 |
0.5 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.1 |
0.3 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 |
0.6 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.1 |
0.3 |
GO:0070101 |
positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.1 |
0.2 |
GO:0045226 |
extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 |
0.3 |
GO:0048749 |
compound eye development(GO:0048749) |
| 0.1 |
0.2 |
GO:0052553 |
induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 |
0.3 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 |
0.2 |
GO:0061534 |
gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 |
0.2 |
GO:0042369 |
vitamin D catabolic process(GO:0042369) |
| 0.1 |
0.5 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 |
0.2 |
GO:1902630 |
regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 |
0.2 |
GO:0098746 |
fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 |
0.2 |
GO:0060979 |
vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.1 |
0.5 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 |
0.6 |
GO:0032926 |
negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 |
0.8 |
GO:0060346 |
bone trabecula formation(GO:0060346) |
| 0.1 |
1.2 |
GO:0016540 |
protein autoprocessing(GO:0016540) |
| 0.1 |
0.2 |
GO:0031630 |
regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.1 |
0.3 |
GO:0060373 |
regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 |
0.4 |
GO:0003150 |
muscular septum morphogenesis(GO:0003150) |
| 0.0 |
0.3 |
GO:0045163 |
clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 |
0.4 |
GO:0038044 |
transforming growth factor-beta secretion(GO:0038044) |
| 0.0 |
0.3 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.0 |
0.2 |
GO:1903566 |
positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 |
0.4 |
GO:1902866 |
regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 |
0.4 |
GO:0000727 |
double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 |
0.1 |
GO:0016539 |
intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 |
0.1 |
GO:2000297 |
negative regulation of synapse maturation(GO:2000297) |
| 0.0 |
0.3 |
GO:0035469 |
determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 |
0.1 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 |
2.5 |
GO:0043486 |
histone exchange(GO:0043486) |
| 0.0 |
0.1 |
GO:0050894 |
determination of affect(GO:0050894) |
| 0.0 |
0.4 |
GO:0045602 |
negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 |
0.1 |
GO:0021816 |
lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 |
0.1 |
GO:0032877 |
positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 |
0.2 |
GO:0061107 |
seminal vesicle development(GO:0061107) |
| 0.0 |
0.1 |
GO:0042938 |
dipeptide transport(GO:0042938) |
| 0.0 |
0.2 |
GO:1903490 |
regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 |
0.4 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.9 |
GO:1990001 |
inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 |
0.2 |
GO:0060748 |
tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 |
0.4 |
GO:0098734 |
macromolecule depalmitoylation(GO:0098734) |
| 0.0 |
0.1 |
GO:0061325 |
cell proliferation involved in outflow tract morphogenesis(GO:0061325) |
| 0.0 |
0.1 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 |
0.3 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 |
0.1 |
GO:1902896 |
terminal web assembly(GO:1902896) |
| 0.0 |
0.6 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.0 |
0.2 |
GO:0051136 |
regulation of NK T cell differentiation(GO:0051136) |
| 0.0 |
0.4 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
0.4 |
GO:0035279 |
mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 |
0.3 |
GO:0045759 |
negative regulation of action potential(GO:0045759) |
| 0.0 |
0.1 |
GO:0019056 |
modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 |
0.3 |
GO:0097646 |
calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 |
0.1 |
GO:0008050 |
female courtship behavior(GO:0008050) |
| 0.0 |
0.3 |
GO:0000055 |
ribosomal large subunit export from nucleus(GO:0000055) ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 |
0.1 |
GO:0038163 |
thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 |
0.2 |
GO:0070120 |
ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 |
0.1 |
GO:0035669 |
TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 |
0.1 |
GO:0021798 |
forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 |
0.1 |
GO:0086097 |
phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 |
0.1 |
GO:0009436 |
glyoxylate catabolic process(GO:0009436) |
| 0.0 |
0.1 |
GO:0046947 |
hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 |
0.1 |
GO:0070295 |
renal water absorption(GO:0070295) |
| 0.0 |
0.1 |
GO:0002296 |
T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 |
0.1 |
GO:2000418 |
positive regulation of eosinophil migration(GO:2000418) |
| 0.0 |
0.1 |
GO:1904954 |
canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.0 |
0.5 |
GO:2000586 |
regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) |
| 0.0 |
0.1 |
GO:1904674 |
positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.0 |
0.3 |
GO:0032074 |
negative regulation of nuclease activity(GO:0032074) |
| 0.0 |
0.1 |
GO:0032487 |
regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 |
0.1 |
GO:0060266 |
negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 |
0.1 |
GO:1905075 |
occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.0 |
0.1 |
GO:2001076 |
regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.0 |
0.2 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 |
0.1 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 |
0.1 |
GO:0051541 |
elastin metabolic process(GO:0051541) |
| 0.0 |
0.4 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.0 |
0.1 |
GO:0060842 |
arterial endothelial cell differentiation(GO:0060842) |
| 0.0 |
0.1 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.0 |
0.0 |
GO:0048936 |
peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 |
0.1 |
GO:2000821 |
regulation of grooming behavior(GO:2000821) |
| 0.0 |
0.7 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.0 |
0.1 |
GO:0034154 |
toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 |
0.2 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 |
0.1 |
GO:1990927 |
negative regulation of synaptic vesicle recycling(GO:1903422) negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 |
0.2 |
GO:0099566 |
regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 |
0.2 |
GO:2000138 |
positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 |
0.1 |
GO:0035021 |
negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 |
0.0 |
GO:2000015 |
regulation of determination of dorsal identity(GO:2000015) |
| 0.0 |
0.3 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.0 |
0.3 |
GO:0015816 |
glycine transport(GO:0015816) |
| 0.0 |
0.5 |
GO:0045187 |
regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 |
0.1 |
GO:0061312 |
BMP signaling pathway involved in heart development(GO:0061312) |
| 0.0 |
0.0 |
GO:0021764 |
amygdala development(GO:0021764) |
| 0.0 |
0.1 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 |
0.4 |
GO:0043517 |
positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 |
0.1 |
GO:0060729 |
amacrine cell differentiation(GO:0035881) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 |
0.2 |
GO:0051388 |
positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 |
0.1 |
GO:0002268 |
follicular dendritic cell differentiation(GO:0002268) |
| 0.0 |
0.2 |
GO:0060316 |
positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 |
0.1 |
GO:0035544 |
negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 |
0.7 |
GO:0007202 |
activation of phospholipase C activity(GO:0007202) |
| 0.0 |
0.1 |
GO:0060282 |
positive regulation of oocyte development(GO:0060282) |
| 0.0 |
0.3 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.0 |
0.2 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.0 |
0.2 |
GO:2000318 |
positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.0 |
0.1 |
GO:0042905 |
9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 |
0.1 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 |
0.0 |
GO:0003241 |
growth involved in heart morphogenesis(GO:0003241) |
| 0.0 |
0.0 |
GO:0032224 |
positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 |
0.0 |
GO:0031548 |
regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 |
0.2 |
GO:0000733 |
DNA strand renaturation(GO:0000733) |
| 0.0 |
0.1 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 |
0.2 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.0 |
0.1 |
GO:0097091 |
synaptic vesicle clustering(GO:0097091) |
| 0.0 |
0.0 |
GO:0003285 |
septum secundum development(GO:0003285) |
| 0.0 |
0.1 |
GO:0070086 |
ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 |
0.1 |
GO:1902748 |
positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 |
0.1 |
GO:0003415 |
chondrocyte hypertrophy(GO:0003415) |
| 0.0 |
0.1 |
GO:0002501 |
peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 |
0.2 |
GO:0051665 |
membrane raft localization(GO:0051665) |
| 0.0 |
0.0 |
GO:0006421 |
asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 |
0.1 |
GO:0060754 |
positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 |
0.1 |
GO:0006226 |
dUMP biosynthetic process(GO:0006226) |
| 0.0 |
0.1 |
GO:0032072 |
plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) regulation of restriction endodeoxyribonuclease activity(GO:0032072) positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 |
0.1 |
GO:0032484 |
Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 |
0.1 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.1 |
GO:0071393 |
cellular response to progesterone stimulus(GO:0071393) |
| 0.0 |
0.0 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) |
| 0.0 |
0.1 |
GO:1902952 |
positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 |
0.1 |
GO:0060290 |
transdifferentiation(GO:0060290) |
| 0.0 |
0.1 |
GO:2000301 |
negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 |
0.1 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 |
0.1 |
GO:0060689 |
cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 |
0.0 |
GO:1901097 |
negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 |
0.1 |
GO:0008588 |
release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 |
0.3 |
GO:0044804 |
nucleophagy(GO:0044804) |
| 0.0 |
0.0 |
GO:0010816 |
substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 |
0.0 |
GO:2001271 |
negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 |
0.0 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 |
0.0 |
GO:1904862 |
inhibitory synapse assembly(GO:1904862) |
| 0.0 |
0.1 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.0 |
0.1 |
GO:1902412 |
regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 |
0.0 |
GO:0044034 |
negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 |
0.0 |
GO:0061314 |
Notch signaling involved in heart development(GO:0061314) |
| 0.0 |
0.2 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.0 |
0.0 |
GO:0061198 |
fungiform papilla formation(GO:0061198) |
| 0.0 |
0.0 |
GO:0051106 |
positive regulation of DNA ligation(GO:0051106) |
| 0.0 |
0.1 |
GO:0007566 |
embryo implantation(GO:0007566) |
| 0.0 |
0.1 |
GO:0060087 |
relaxation of vascular smooth muscle(GO:0060087) |