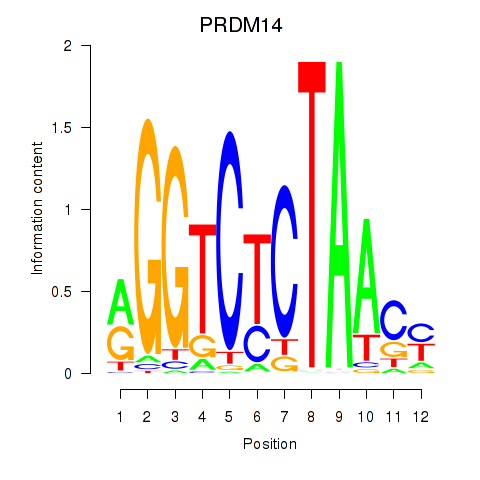
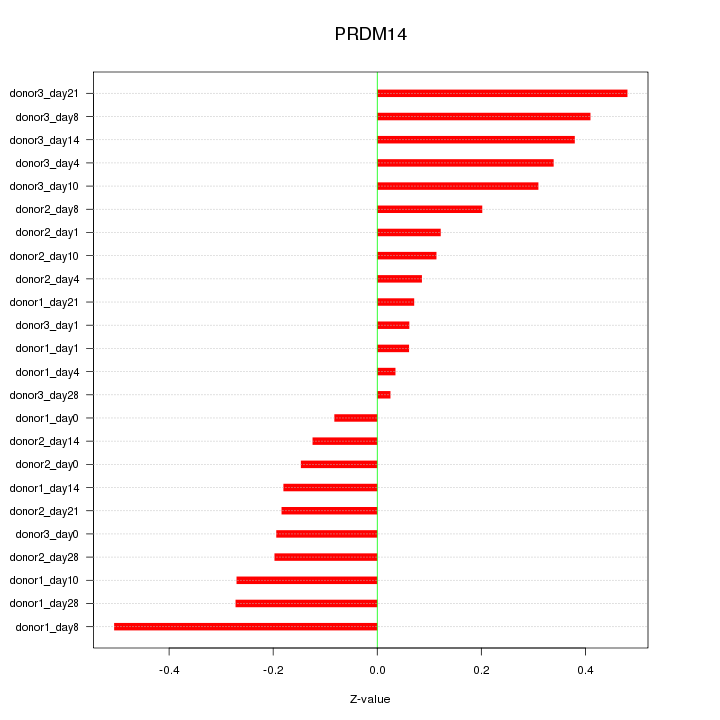
|
chr16_-_55866997
|
0.443
|
ENST00000360526.3
ENST00000361503.4
|
CES1
|
carboxylesterase 1
|
|
chr4_-_103266626
|
0.195
|
ENST00000356736.4
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8
|
|
chr2_+_101437487
|
0.149
|
ENST00000427413.1
ENST00000542504.1
|
NPAS2
|
neuronal PAS domain protein 2
|
|
chr2_-_69098566
|
0.148
|
ENST00000295379.1
|
BMP10
|
bone morphogenetic protein 10
|
|
chr19_+_49838653
|
0.134
|
ENST00000598095.1
ENST00000426897.2
ENST00000323906.4
ENST00000535669.2
ENST00000597602.1
ENST00000595660.1
|
CD37
|
CD37 molecule
|
|
chr12_-_49351228
|
0.116
|
ENST00000541959.1
ENST00000447318.2
|
ARF3
|
ADP-ribosylation factor 3
|
|
chrX_+_41193407
|
0.115
|
ENST00000457138.2
ENST00000441189.2
|
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked
|
|
chr12_-_49351148
|
0.093
|
ENST00000539611.1
ENST00000398092.4
|
ARF3
RP11-302B13.5
|
ADP-ribosylation factor 3
ADP-ribosylation factor 3
|
|
chr17_-_46691990
|
0.088
|
ENST00000576562.1
|
HOXB8
|
homeobox B8
|
|
chrX_+_15525426
|
0.088
|
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr7_-_139763521
|
0.083
|
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12
|
|
chr12_-_49351303
|
0.083
|
ENST00000256682.4
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr6_-_46048116
|
0.080
|
ENST00000185206.6
|
CLIC5
|
chloride intracellular channel 5
|
|
chr17_+_41924511
|
0.075
|
ENST00000377203.4
ENST00000539718.1
ENST00000588884.1
ENST00000293396.8
ENST00000586233.1
|
CD300LG
|
CD300 molecule-like family member g
|
|
chr12_+_56325812
|
0.075
|
ENST00000394147.1
ENST00000551156.1
ENST00000553783.1
ENST00000557080.1
ENST00000432422.3
ENST00000556001.1
|
DGKA
|
diacylglycerol kinase, alpha 80kDa
|
|
chr9_+_132099158
|
0.074
|
ENST00000444125.1
|
RP11-65J3.1
|
RP11-65J3.1
|
|
chr20_-_3154162
|
0.073
|
ENST00000360342.3
|
LZTS3
|
Homo sapiens leucine zipper, putative tumor suppressor family member 3 (LZTS3), mRNA.
|
|
chr17_-_46692287
|
0.072
|
ENST00000239144.4
|
HOXB8
|
homeobox B8
|
|
chrX_+_119292467
|
0.071
|
ENST00000371388.3
|
RHOXF2
|
Rhox homeobox family, member 2
|
|
chr1_+_36621529
|
0.071
|
ENST00000316156.4
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr9_+_132597722
|
0.070
|
ENST00000372429.3
ENST00000315480.4
ENST00000358355.1
|
USP20
|
ubiquitin specific peptidase 20
|
|
chr7_-_124405681
|
0.070
|
ENST00000303921.2
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chr19_+_3572758
|
0.068
|
ENST00000416526.1
|
HMG20B
|
high mobility group 20B
|
|
chr14_+_89290965
|
0.068
|
ENST00000345383.5
ENST00000536576.1
ENST00000346301.4
ENST00000338104.6
ENST00000354441.6
ENST00000380656.2
ENST00000556651.1
ENST00000554686.1
|
TTC8
|
tetratricopeptide repeat domain 8
|
|
chr22_-_29457832
|
0.068
|
ENST00000216071.4
|
C22orf31
|
chromosome 22 open reading frame 31
|
|
chr6_+_72926145
|
0.060
|
ENST00000425662.2
ENST00000453976.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr7_+_150211918
|
0.057
|
ENST00000313543.4
|
GIMAP7
|
GTPase, IMAP family member 7
|
|
chr5_+_32710736
|
0.055
|
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr2_-_228244013
|
0.055
|
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20
|
|
chr17_+_39994032
|
0.052
|
ENST00000293303.4
ENST00000438813.1
|
KLHL10
|
kelch-like family member 10
|
|
chr17_+_47653178
|
0.051
|
ENST00000328741.5
|
NXPH3
|
neurexophilin 3
|
|
chr5_-_108063949
|
0.051
|
ENST00000606054.1
|
LINC01023
|
long intergenic non-protein coding RNA 1023
|
|
chr3_-_155394152
|
0.049
|
ENST00000494598.1
|
PLCH1
|
phospholipase C, eta 1
|
|
chr8_+_110346546
|
0.047
|
ENST00000521662.1
ENST00000521688.1
ENST00000520147.1
|
ENY2
|
enhancer of yellow 2 homolog (Drosophila)
|
|
chr3_-_143567262
|
0.047
|
ENST00000474151.1
ENST00000316549.6
|
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9
|
|
chr2_-_192711968
|
0.046
|
ENST00000304141.4
|
SDPR
|
serum deprivation response
|
|
chr5_+_32711419
|
0.046
|
ENST00000265074.8
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr17_+_36873677
|
0.045
|
ENST00000471200.1
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr10_+_12171636
|
0.044
|
ENST00000379051.1
ENST00000379033.3
ENST00000441368.1
ENST00000298428.9
ENST00000304267.8
|
SEC61A2
|
Sec61 alpha 2 subunit (S. cerevisiae)
|
|
chr15_+_63188009
|
0.041
|
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2
|
|
chr11_-_65625014
|
0.041
|
ENST00000534784.1
|
CFL1
|
cofilin 1 (non-muscle)
|
|
chr19_+_39109735
|
0.040
|
ENST00000593149.1
ENST00000248342.4
ENST00000538434.1
ENST00000588934.1
ENST00000545173.2
ENST00000589307.1
ENST00000586513.1
ENST00000591409.1
ENST00000592558.1
|
EIF3K
|
eukaryotic translation initiation factor 3, subunit K
|
|
chr3_+_10289707
|
0.038
|
ENST00000287652.4
|
TATDN2
|
TatD DNase domain containing 2
|
|
chr10_+_12171721
|
0.038
|
ENST00000379020.4
ENST00000379017.3
|
SEC61A2
|
Sec61 alpha 2 subunit (S. cerevisiae)
|
|
chr19_-_53757941
|
0.037
|
ENST00000594517.1
ENST00000601413.1
ENST00000594681.1
|
ZNF677
|
zinc finger protein 677
|
|
chr11_-_65624415
|
0.036
|
ENST00000524553.1
ENST00000527344.1
|
CFL1
|
cofilin 1 (non-muscle)
|
|
chr7_-_44580861
|
0.033
|
ENST00000546276.1
ENST00000289547.4
ENST00000381160.3
ENST00000423141.1
|
NPC1L1
|
NPC1-like 1
|
|
chr4_+_4861385
|
0.033
|
ENST00000382723.4
|
MSX1
|
msh homeobox 1
|
|
chr3_-_47517302
|
0.033
|
ENST00000441517.2
ENST00000545718.1
|
SCAP
|
SREBF chaperone
|
|
chr1_+_50571949
|
0.028
|
ENST00000357083.4
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4
|
|
chr1_-_27216729
|
0.027
|
ENST00000431781.2
ENST00000374135.4
|
GPN2
|
GPN-loop GTPase 2
|
|
chr17_+_75450075
|
0.027
|
ENST00000592951.1
|
SEPT9
|
septin 9
|
|
chr1_+_155006300
|
0.025
|
ENST00000295542.1
ENST00000392480.1
ENST00000423025.2
ENST00000368419.2
|
DCST1
|
DC-STAMP domain containing 1
|
|
chr20_-_48770174
|
0.025
|
ENST00000341698.2
|
TMEM189-UBE2V1
|
TMEM189-UBE2V1 readthrough
|
|
chr11_-_46615498
|
0.024
|
ENST00000533727.1
ENST00000534300.1
ENST00000528950.1
ENST00000526606.1
|
AMBRA1
|
autophagy/beclin-1 regulator 1
|
|
chr5_-_16936340
|
0.023
|
ENST00000507288.1
ENST00000513610.1
|
MYO10
|
myosin X
|
|
chr17_-_73775839
|
0.021
|
ENST00000592643.1
ENST00000591890.1
ENST00000587171.1
ENST00000254810.4
ENST00000589599.1
|
H3F3B
|
H3 histone, family 3B (H3.3B)
|
|
chr11_+_85566422
|
0.021
|
ENST00000342404.3
|
CCDC83
|
coiled-coil domain containing 83
|
|
chr17_-_19651654
|
0.021
|
ENST00000395555.3
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1
|
|
chr18_+_57567180
|
0.019
|
ENST00000316660.6
ENST00000269518.9
|
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1
|
|
chr2_-_208030647
|
0.014
|
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr7_-_30066233
|
0.014
|
ENST00000222803.5
|
FKBP14
|
FK506 binding protein 14, 22 kDa
|
|
chr8_-_102216925
|
0.014
|
ENST00000517844.1
|
ZNF706
|
zinc finger protein 706
|
|
chr11_+_62475130
|
0.012
|
ENST00000294117.5
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3
|
|
chr12_-_52967600
|
0.012
|
ENST00000549343.1
ENST00000305620.2
|
KRT74
|
keratin 74
|
|
chr15_+_69850521
|
0.012
|
ENST00000558781.1
|
RP11-279F6.1
|
RP11-279F6.1
|
|
chr2_-_145277882
|
0.012
|
ENST00000465070.1
ENST00000444559.1
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr1_+_156052354
|
0.011
|
ENST00000368301.2
|
LMNA
|
lamin A/C
|
|
chr11_-_105892937
|
0.011
|
ENST00000301919.4
ENST00000534458.1
ENST00000530108.1
ENST00000530788.1
|
MSANTD4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils
|
|
chr17_-_19651668
|
0.010
|
ENST00000494157.2
ENST00000225740.6
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1
|
|
chr1_-_116383322
|
0.007
|
ENST00000429731.1
|
NHLH2
|
nescient helix loop helix 2
|
|
chr19_-_6279932
|
0.006
|
ENST00000252674.7
|
MLLT1
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1
|
|
chr17_-_17485731
|
0.005
|
ENST00000395783.1
|
PEMT
|
phosphatidylethanolamine N-methyltransferase
|
|
chr1_-_31845914
|
0.003
|
ENST00000373713.2
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor)
|
|
chr4_+_81187753
|
0.003
|
ENST00000312465.7
ENST00000456523.3
|
FGF5
|
fibroblast growth factor 5
|
|
chr17_+_41924536
|
0.002
|
ENST00000317310.4
|
CD300LG
|
CD300 molecule-like family member g
|
|
chr19_-_49314269
|
0.001
|
ENST00000545387.2
ENST00000316273.6
ENST00000402551.1
ENST00000598162.1
ENST00000599246.1
|
BCAT2
|
branched chain amino-acid transaminase 2, mitochondrial
|
|
chr3_+_155838337
|
0.001
|
ENST00000490337.1
ENST00000389636.5
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr14_-_21492251
|
0.001
|
ENST00000554398.1
|
NDRG2
|
NDRG family member 2
|
|
chr21_+_26934165
|
0.000
|
ENST00000456917.1
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chr14_-_21492113
|
0.000
|
ENST00000554094.1
|
NDRG2
|
NDRG family member 2
|