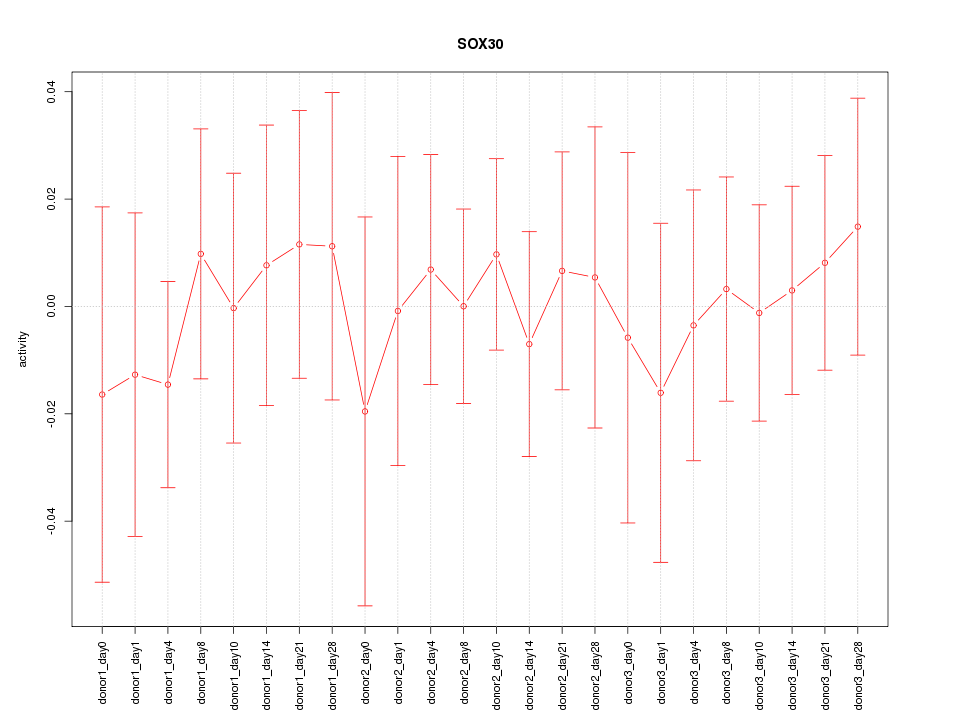
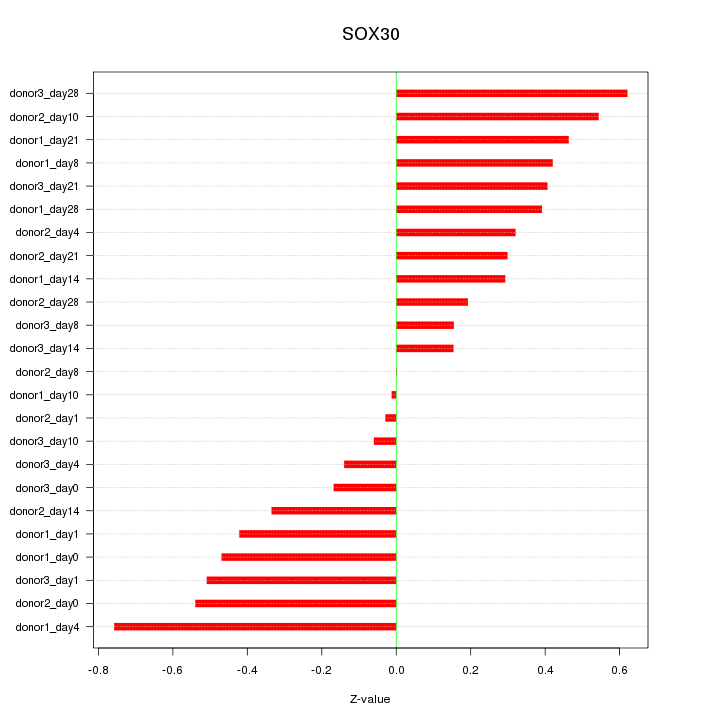
Motif ID: SOX30
Z-value: 0.379
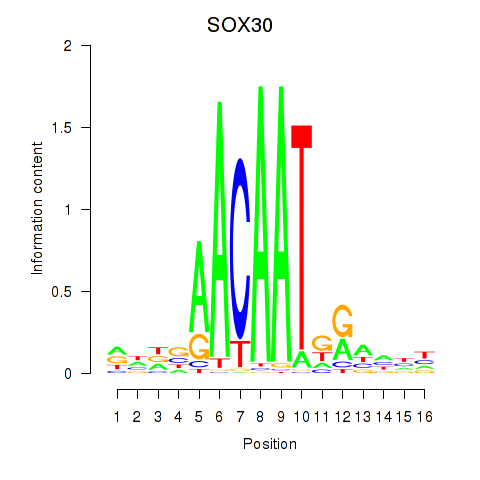
Transcription factors associated with SOX30:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| SOX30 | ENSG00000039600.6 | SOX30 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX30 | hg19_v2_chr5_-_157079428_157079452, hg19_v2_chr5_-_157079372_157079395 | -0.21 | 3.2e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.2 | 0.6 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.1 | 0.4 | GO:0034239 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.1 | 1.5 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.1 | 0.3 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 1.6 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 0.6 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.1 | 0.8 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.2 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.4 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.4 | GO:0048505 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.1 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.4 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.0 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.0 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.1 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.4 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.6 | GO:0050294 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 1.6 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 1.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME_CYTOSOLIC_SULFONATION_OF_SMALL_MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |