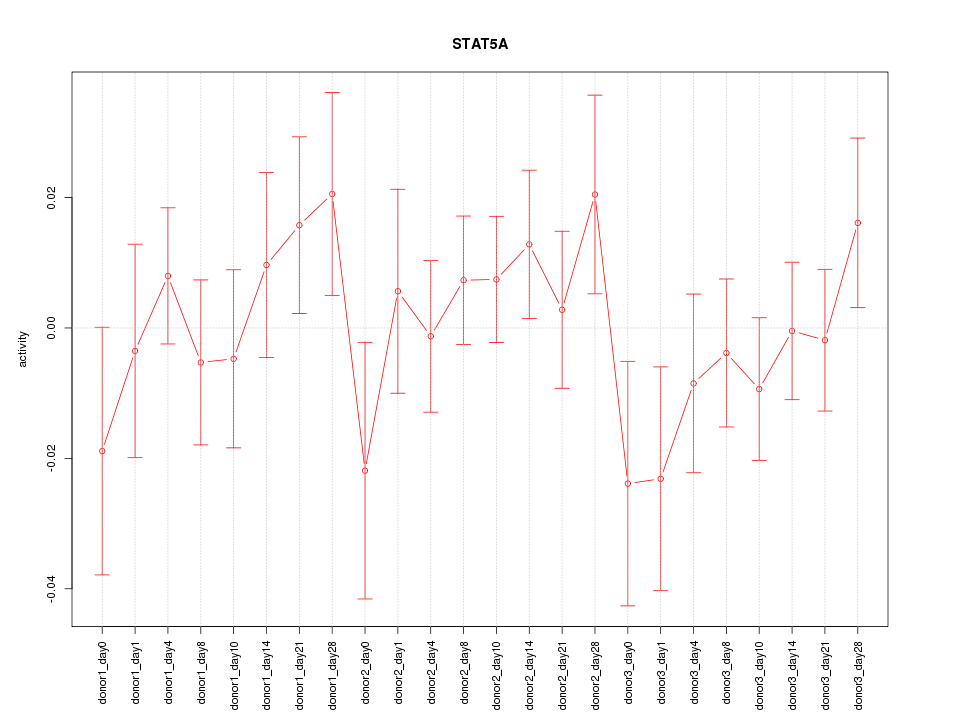
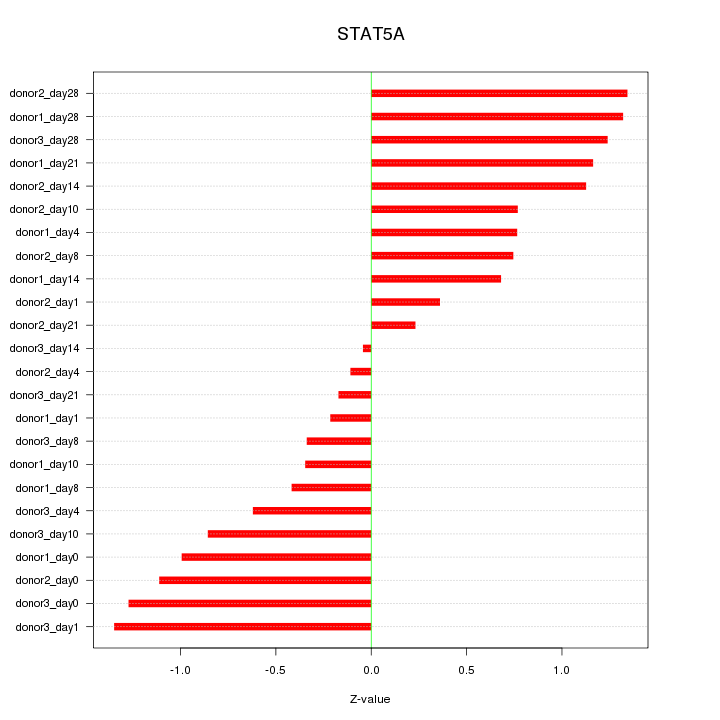
Motif ID: STAT5A
Z-value: 0.851
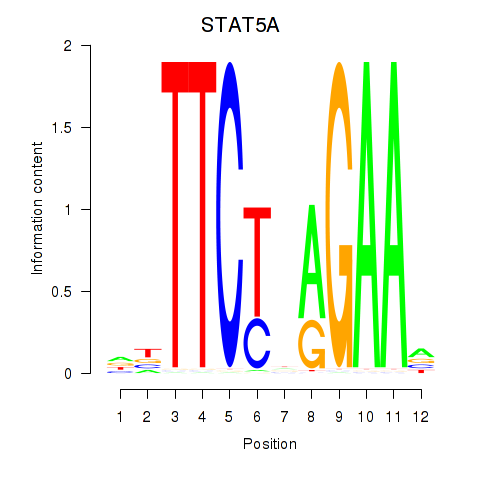
Transcription factors associated with STAT5A:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| STAT5A | ENSG00000126561.12 | STAT5A |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT5A | hg19_v2_chr17_+_40440481_40440561 | 0.73 | 5.4e-05 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.6 | 2.4 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.5 | 2.2 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.5 | 1.6 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.4 | 2.8 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.4 | 1.2 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.4 | 1.6 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.4 | 1.1 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.4 | 2.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.3 | 1.0 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.3 | 2.3 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.3 | 2.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.3 | 0.9 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.3 | 1.7 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.3 | 0.8 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.3 | 1.3 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.3 | 1.0 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.3 | 1.0 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.2 | 1.0 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.2 | 1.0 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.2 | 0.9 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.2 | 0.9 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.2 | 0.9 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.2 | 2.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 6.9 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 7.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 1.0 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.2 | 1.2 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.2 | 0.6 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.2 | 0.9 | GO:0072313 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.2 | 1.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 0.9 | GO:0034486 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) |
| 0.2 | 0.5 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.2 | 0.5 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.2 | 0.7 | GO:0090264 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) response to high density lipoprotein particle(GO:0055099) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) |
| 0.2 | 2.9 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.2 | 2.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 1.5 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.2 | 0.5 | GO:1902161 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.2 | 0.8 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.2 | 0.6 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 0.8 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.4 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.1 | 0.3 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.4 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.7 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 | 1.0 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 1.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 0.4 | GO:0001983 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.1 | 1.0 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.4 | GO:0070428 | negative regulation of interleukin-23 production(GO:0032707) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.1 | 0.3 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.1 | 0.3 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.1 | 0.3 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 0.3 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 0.3 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.1 | 1.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 2.0 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.3 | GO:0090024 | complement component C5a signaling pathway(GO:0038178) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.3 | GO:1902824 | positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.1 | 0.4 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.3 | GO:1902995 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.1 | 0.2 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 1.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.6 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 1.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 2.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.8 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 1.4 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.2 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 0.2 | GO:1902996 | regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.1 | 0.3 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 | 0.2 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.1 | 0.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.0 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 0.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.2 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.4 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 0.1 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.1 | 0.1 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.1 | 0.3 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 0.6 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.7 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.5 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.3 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.1 | 0.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.9 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 0.2 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 0.2 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.1 | 0.2 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.1 | 0.1 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.1 | 0.2 | GO:1990641 | cellular response to bile acid(GO:1903413) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.7 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 0.6 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 0.2 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.1 | 0.4 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.1 | 2.4 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 0.8 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.1 | 0.2 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 0.3 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.1 | 0.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.3 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.8 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.3 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.1 | 0.1 | GO:0098743 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.1 | 0.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.2 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.1 | 0.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.2 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.3 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.1 | 0.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.4 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.0 | 0.1 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.6 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.8 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 1.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.5 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.0 | 0.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.0 | 0.1 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.5 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.3 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.4 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.4 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 1.0 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.3 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0030473 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.2 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.2 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.3 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.2 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 0.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.6 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.3 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 1.7 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.5 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.2 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 0.3 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.0 | 0.6 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 1.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 1.4 | GO:0043030 | regulation of macrophage activation(GO:0043030) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.6 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.3 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.1 | GO:0090427 | monocyte extravasation(GO:0035696) regulation of fertilization(GO:0080154) activation of meiosis(GO:0090427) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 2.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 2.5 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.5 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.4 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.6 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.3 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.3 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 1.8 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.6 | GO:0002320 | lymphoid progenitor cell differentiation(GO:0002320) |
| 0.0 | 0.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.2 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.2 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.3 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.6 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.6 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 5.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.3 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.2 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.3 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 3.1 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.5 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.4 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.2 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 1.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.2 | GO:0001956 | positive regulation of neurotransmitter secretion(GO:0001956) |
| 0.0 | 0.1 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 1.0 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 1.3 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 0.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 1.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.2 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.0 | 0.6 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.3 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.0 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 0.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.5 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:0071351 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.4 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.3 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.2 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.4 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.1 | GO:0002418 | immune response to tumor cell(GO:0002418) |
| 0.0 | 1.1 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 1.5 | GO:0001942 | hair follicle development(GO:0001942) |
| 0.0 | 0.1 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.0 | 0.1 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.6 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.2 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.3 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.3 | GO:0051168 | nuclear export(GO:0051168) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.0 | GO:0036302 | atrioventricular canal development(GO:0036302) |
| 0.0 | 0.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.3 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 2.9 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.1 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.3 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.0 | GO:0014744 | positive regulation of muscle adaptation(GO:0014744) |
| 0.0 | 0.7 | GO:1903901 | negative regulation of viral process(GO:0048525) negative regulation of viral life cycle(GO:1903901) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.2 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.4 | 0.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.4 | 1.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.3 | 1.6 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.3 | 0.9 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.3 | 6.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 0.8 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.2 | 0.6 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.2 | 0.6 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.9 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.1 | 1.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 3.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.5 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 1.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.7 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 0.5 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 0.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.5 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 1.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 1.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.7 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 2.2 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.5 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.3 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 1.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.7 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.4 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.8 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.2 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 0.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.2 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.9 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.5 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.3 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.8 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 3.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.0 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 1.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.3 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 1.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.3 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 1.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.4 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.5 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.2 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 2.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 2.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 1.0 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 1.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.3 | GO:0042629 | mast cell granule(GO:0042629) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.6 | 2.3 | GO:1904493 | Ac-Asp-Glu binding(GO:1904492) tetrahydrofolyl-poly(glutamate) polymer binding(GO:1904493) |
| 0.5 | 1.6 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.4 | 7.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.4 | 5.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.3 | 2.4 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.3 | 1.0 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.3 | 1.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.3 | 1.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.3 | 0.8 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.3 | 1.9 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.3 | 1.0 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.3 | 1.0 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.2 | 1.4 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.2 | 0.9 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 3.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 0.8 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 2.7 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.2 | 1.9 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 1.2 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.2 | 0.6 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.2 | 0.8 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.2 | 2.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 2.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 0.5 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.2 | 0.6 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.1 | 2.0 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.6 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.1 | 0.4 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 2.9 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.6 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.1 | 1.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.3 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.1 | 0.4 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.1 | 0.3 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 1.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 1.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.6 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.9 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 1.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.3 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 1.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.0 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1.0 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.5 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.3 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 1.0 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 1.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.3 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.3 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 1.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.2 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.1 | 0.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 0.4 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.4 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 0.2 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 0.4 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.8 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.1 | 0.4 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.2 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 0.2 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.1 | 0.2 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.1 | 0.2 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.1 | 0.2 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.1 | 2.2 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 1.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.8 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.3 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.1 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.3 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.1 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 1.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.5 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.4 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 1.0 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.3 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 1.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 3.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 1.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.4 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 1.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 2.9 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.3 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.5 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.5 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.3 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.2 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.6 | GO:0034061 | DNA polymerase activity(GO:0034061) |
| 0.0 | 0.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 1.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.0 | GO:0052726 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.0 | 0.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 1.0 | GO:0016279 | protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.2 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.0 | 2.3 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.9 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 3.6 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID_VEGFR1_2_PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.1 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | ST_TYPE_I_INTERFERON_PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.4 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.5 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 4.7 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.0 | PID_CERAMIDE_PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.6 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | PID_PDGFRA_PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.0 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.5 | PID_CD8_TCR_DOWNSTREAM_PATHWAY | Downstream signaling in naïve CD8+ T cells |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_24_HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.6 | REACTOME_PROSTANOID_LIGAND_RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 1.9 | REACTOME_RECYCLING_OF_BILE_ACIDS_AND_SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 2.7 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 2.4 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.6 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.2 | REACTOME_PURINE_CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 0.7 | REACTOME_APOBEC3G_MEDIATED_RESISTANCE_TO_HIV1_INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 1.1 | REACTOME_REVERSIBLE_HYDRATION_OF_CARBON_DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 0.9 | REACTOME_ETHANOL_OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 0.8 | REACTOME_CREATION_OF_C4_AND_C2_ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.8 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.8 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.0 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.8 | REACTOME_ENDOGENOUS_STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.6 | REACTOME_INHIBITION_OF_REPLICATION_INITIATION_OF_DAMAGED_DNA_BY_RB1_E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 1.0 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_GOLGI_MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.0 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.1 | REACTOME_PEROXISOMAL_LIPID_METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.1 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.6 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.0 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.4 | REACTOME_ELEVATION_OF_CYTOSOLIC_CA2_LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.0 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.2 | REACTOME_LYSOSOME_VESICLE_BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME_BETA_DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.5 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.6 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 2.1 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.6 | REACTOME_SHC_MEDIATED_SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.6 | REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.5 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.7 | REACTOME_INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.3 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.8 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME_PHOSPHORYLATION_OF_THE_APC_C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.9 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.6 | REACTOME_COMPLEMENT_CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.6 | REACTOME_RNA_POL_I_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.8 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME_REGULATION_OF_KIT_SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.6 | REACTOME_CYTOCHROME_P450_ARRANGED_BY_SUBSTRATE_TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.9 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME_NFKB_ACTIVATION_THROUGH_FADD_RIP1_PATHWAY_MEDIATED_BY_CASPASE_8_AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.2 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.5 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.1 | REACTOME_ANTIGEN_PROCESSING_CROSS_PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.2 | REACTOME_NONSENSE_MEDIATED_DECAY_ENHANCED_BY_THE_EXON_JUNCTION_COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.3 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |