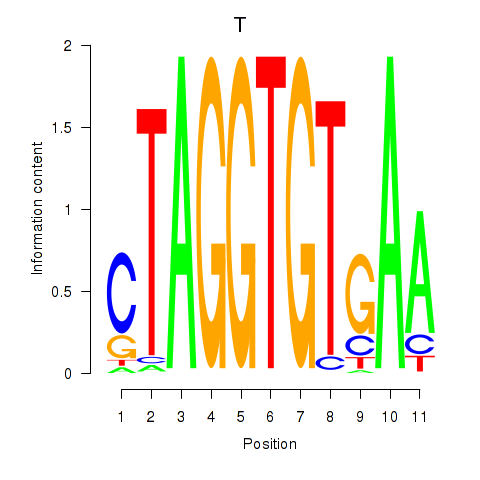
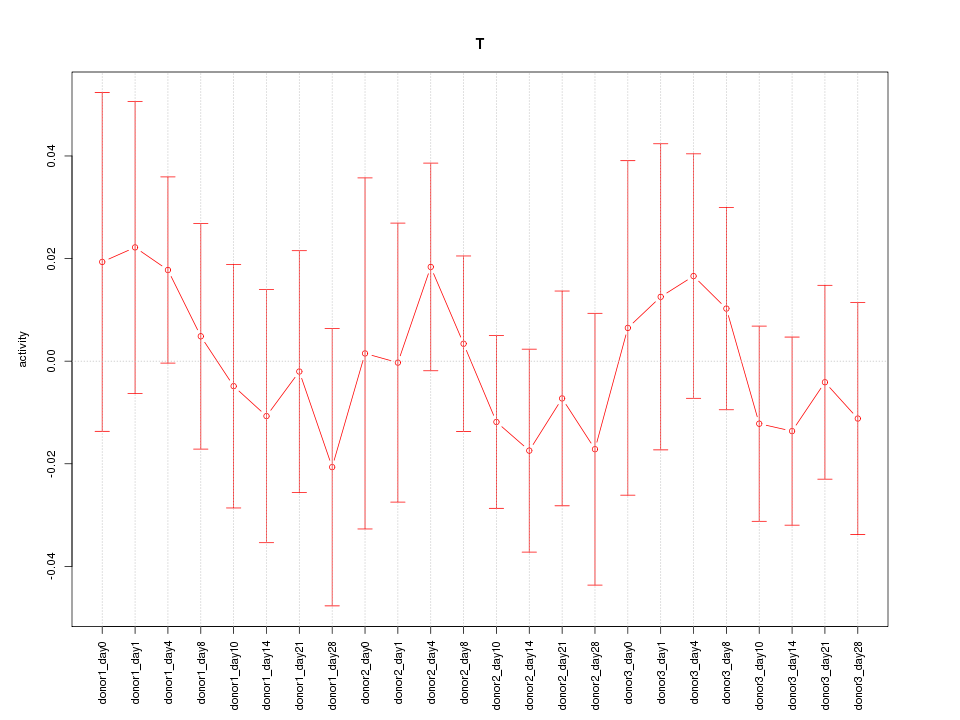
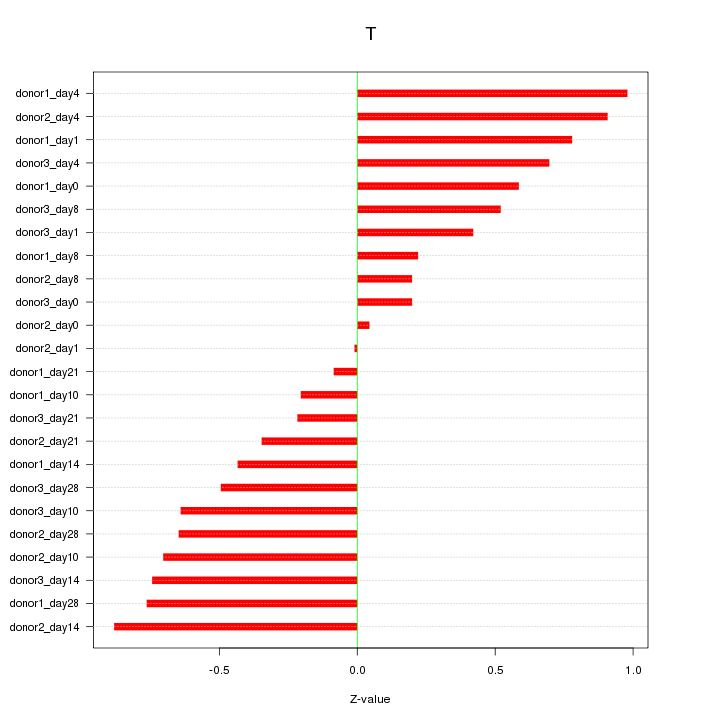
| 0.3 |
1.4 |
GO:0016103 |
diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.3 |
1.3 |
GO:0038195 |
urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.2 |
0.9 |
GO:0035627 |
ceramide transport(GO:0035627) |
| 0.2 |
1.5 |
GO:0001661 |
conditioned taste aversion(GO:0001661) |
| 0.1 |
0.7 |
GO:0002669 |
positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) protein K29-linked ubiquitination(GO:0035519) |
| 0.1 |
1.0 |
GO:0002318 |
myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 |
0.6 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 |
0.6 |
GO:2001268 |
negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.1 |
0.6 |
GO:0035803 |
egg coat formation(GO:0035803) |
| 0.1 |
0.3 |
GO:1900108 |
negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 |
0.7 |
GO:0009753 |
response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 |
0.3 |
GO:0045210 |
negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
| 0.1 |
0.5 |
GO:0002018 |
renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.1 |
0.2 |
GO:0048633 |
positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 |
1.0 |
GO:0019388 |
galactose catabolic process(GO:0019388) |
| 0.0 |
0.3 |
GO:1902963 |
regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 |
0.3 |
GO:0030805 |
regulation of cyclic nucleotide catabolic process(GO:0030805) regulation of cAMP catabolic process(GO:0030820) regulation of purine nucleotide catabolic process(GO:0033121) regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 |
0.2 |
GO:0050668 |
cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.0 |
0.2 |
GO:0003241 |
growth involved in heart morphogenesis(GO:0003241) |
| 0.0 |
1.4 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
1.1 |
GO:0006346 |
methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 |
3.1 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.0 |
0.4 |
GO:2000049 |
positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 |
0.5 |
GO:0052696 |
flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 |
1.3 |
GO:0007398 |
ectoderm development(GO:0007398) |
| 0.0 |
0.1 |
GO:1901069 |
guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 |
0.8 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 |
0.1 |
GO:0060492 |
foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 |
0.1 |
GO:0039019 |
pronephric nephron development(GO:0039019) |
| 0.0 |
0.3 |
GO:0070973 |
protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 |
0.6 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.0 |
0.2 |
GO:0070537 |
histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 |
0.1 |
GO:0098759 |
response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.0 |
0.3 |
GO:0033601 |
positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 |
0.1 |
GO:0001757 |
somite specification(GO:0001757) |
| 0.0 |
0.3 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.0 |
0.2 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 |
0.1 |
GO:0001207 |
histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 |
0.2 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 |
0.2 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 |
0.1 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 |
0.1 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 |
0.1 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 |
0.1 |
GO:2000158 |
positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 |
1.2 |
GO:0000186 |
activation of MAPKK activity(GO:0000186) |
| 0.0 |
0.2 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 |
0.3 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 |
0.1 |
GO:0033686 |
positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 |
0.2 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.0 |
0.1 |
GO:2000490 |
negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 |
0.1 |
GO:0048539 |
bone marrow development(GO:0048539) |
| 0.0 |
1.0 |
GO:0008631 |
intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.0 |
0.4 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 |
0.1 |
GO:1905150 |
regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 |
0.5 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 |
0.1 |
GO:0043461 |
proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 |
0.3 |
GO:0060445 |
branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 |
0.2 |
GO:0098779 |
mitophagy in response to mitochondrial depolarization(GO:0098779) |
| 0.0 |
0.5 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.5 |
GO:0045104 |
intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 |
0.2 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.1 |
GO:0045653 |
negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 |
0.2 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |