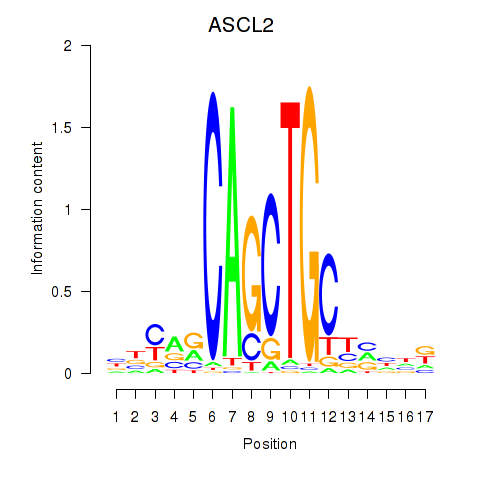
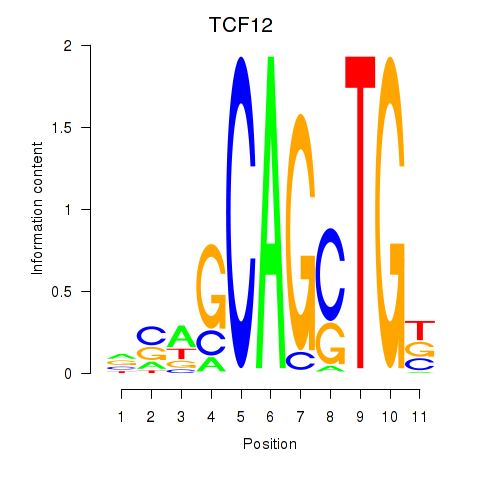
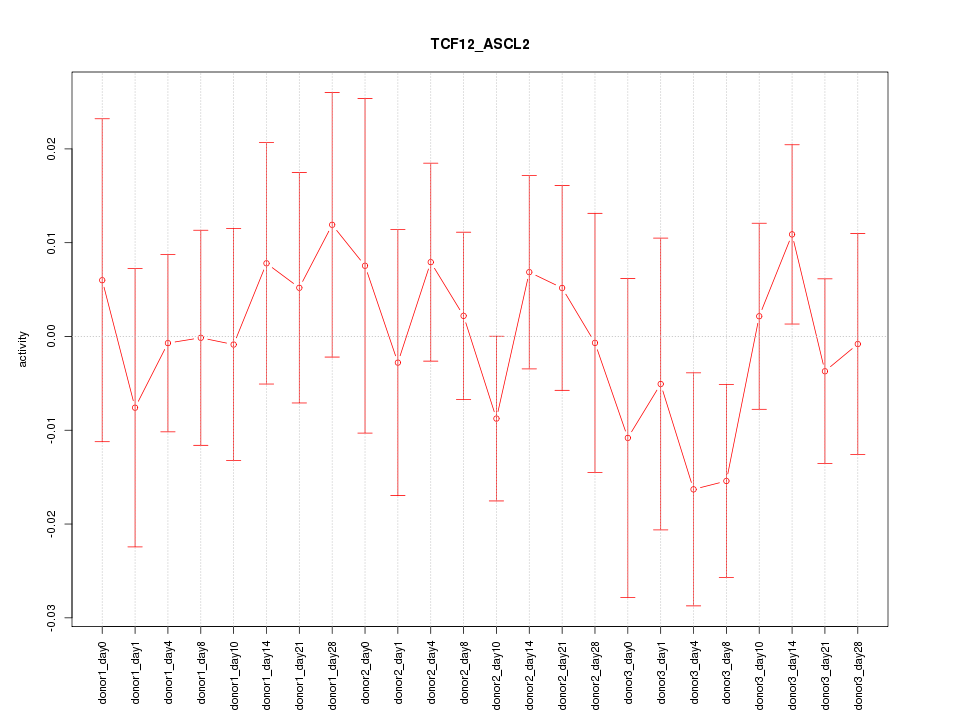
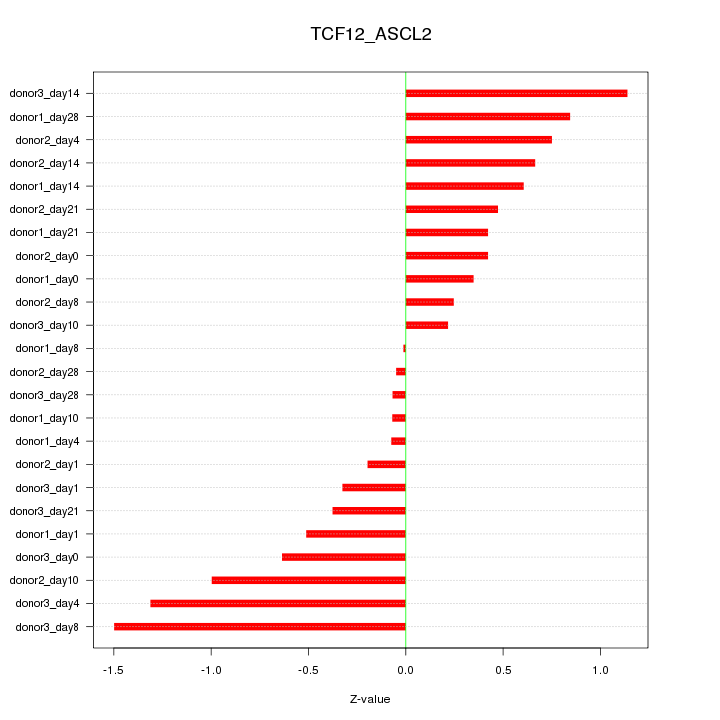
| 0.7 |
2.8 |
GO:2000768 |
glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.4 |
1.4 |
GO:0097089 |
methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.3 |
1.9 |
GO:0050915 |
sensory perception of sour taste(GO:0050915) |
| 0.2 |
0.7 |
GO:1904899 |
regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.2 |
0.8 |
GO:0033634 |
positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.2 |
0.5 |
GO:0009258 |
10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.2 |
0.8 |
GO:0015670 |
carbon dioxide transport(GO:0015670) |
| 0.2 |
0.7 |
GO:0005986 |
sucrose biosynthetic process(GO:0005986) |
| 0.2 |
0.5 |
GO:0021571 |
rhombomere 5 development(GO:0021571) |
| 0.1 |
0.1 |
GO:0040037 |
negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.1 |
0.5 |
GO:0048389 |
intermediate mesoderm development(GO:0048389) pattern specification involved in mesonephros development(GO:0061227) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.1 |
0.4 |
GO:0006667 |
sphinganine metabolic process(GO:0006667) |
| 0.1 |
0.1 |
GO:0032878 |
regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 |
0.4 |
GO:0070105 |
positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 |
0.4 |
GO:0055073 |
cadmium ion homeostasis(GO:0055073) lysosomal membrane organization(GO:0097212) negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.1 |
0.8 |
GO:0098704 |
fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.1 |
1.0 |
GO:0048630 |
skeletal muscle tissue growth(GO:0048630) |
| 0.1 |
0.9 |
GO:0043353 |
enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 |
0.3 |
GO:0090076 |
relaxation of skeletal muscle(GO:0090076) |
| 0.1 |
0.3 |
GO:1902309 |
negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 |
0.4 |
GO:0032972 |
regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 |
0.4 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.1 |
0.1 |
GO:0072229 |
proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.1 |
0.4 |
GO:0018032 |
protein amidation(GO:0018032) |
| 0.1 |
0.7 |
GO:0009804 |
coumarin metabolic process(GO:0009804) |
| 0.1 |
0.4 |
GO:0060708 |
spongiotrophoblast differentiation(GO:0060708) |
| 0.1 |
0.7 |
GO:0098535 |
de novo centriole assembly(GO:0098535) |
| 0.1 |
0.3 |
GO:0032687 |
negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 |
0.3 |
GO:0033214 |
iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 0.1 |
0.3 |
GO:0003285 |
septum secundum development(GO:0003285) |
| 0.1 |
0.7 |
GO:0021678 |
third ventricle development(GO:0021678) |
| 0.1 |
0.4 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.1 |
0.3 |
GO:0021966 |
corticospinal neuron axon guidance(GO:0021966) |
| 0.1 |
0.2 |
GO:0072061 |
inner medullary collecting duct development(GO:0072061) |
| 0.1 |
0.3 |
GO:0009386 |
translational attenuation(GO:0009386) |
| 0.1 |
0.3 |
GO:0090299 |
regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.1 |
0.7 |
GO:0051388 |
positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 |
0.2 |
GO:0021524 |
visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) cardiac cell fate determination(GO:0060913) |
| 0.1 |
0.4 |
GO:0034154 |
toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 |
0.2 |
GO:0033591 |
response to L-ascorbic acid(GO:0033591) |
| 0.1 |
0.4 |
GO:0001705 |
ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.1 |
0.2 |
GO:0038109 |
response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.1 |
0.1 |
GO:0060711 |
labyrinthine layer development(GO:0060711) |
| 0.1 |
0.3 |
GO:0050916 |
sensory perception of sweet taste(GO:0050916) |
| 0.1 |
0.2 |
GO:0006258 |
UDP-glucose catabolic process(GO:0006258) |
| 0.1 |
0.7 |
GO:1903273 |
regulation of sodium ion export(GO:1903273) positive regulation of sodium ion export(GO:1903275) regulation of sodium ion export from cell(GO:1903276) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.1 |
0.7 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.1 |
0.2 |
GO:0060129 |
thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.1 |
0.5 |
GO:1990504 |
dense core granule exocytosis(GO:1990504) |
| 0.1 |
0.6 |
GO:0098914 |
membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 |
0.5 |
GO:0016191 |
synaptic vesicle uncoating(GO:0016191) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 |
0.2 |
GO:0045976 |
negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.1 |
0.2 |
GO:0072134 |
nephrogenic mesenchyme morphogenesis(GO:0072134) allantois development(GO:1905069) |
| 0.1 |
0.7 |
GO:0006228 |
UTP biosynthetic process(GO:0006228) |
| 0.1 |
0.1 |
GO:0072199 |
mesenchymal cell proliferation involved in ureter development(GO:0072198) regulation of mesenchymal cell proliferation involved in ureter development(GO:0072199) |
| 0.1 |
0.8 |
GO:0036159 |
inner dynein arm assembly(GO:0036159) |
| 0.1 |
0.6 |
GO:0043152 |
induction of bacterial agglutination(GO:0043152) |
| 0.1 |
0.2 |
GO:0006059 |
hexitol metabolic process(GO:0006059) |
| 0.1 |
0.8 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.1 |
0.2 |
GO:0032304 |
negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 |
0.3 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 |
0.2 |
GO:0098937 |
dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.1 |
0.3 |
GO:2001293 |
malonyl-CoA metabolic process(GO:2001293) |
| 0.1 |
0.2 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 |
0.2 |
GO:0031508 |
pericentric heterochromatin assembly(GO:0031508) |
| 0.1 |
0.3 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 |
0.5 |
GO:0003356 |
regulation of cilium beat frequency(GO:0003356) |
| 0.0 |
0.3 |
GO:0015798 |
myo-inositol transport(GO:0015798) |
| 0.0 |
1.5 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 |
0.0 |
GO:1901187 |
regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 |
0.2 |
GO:0007228 |
positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 |
0.4 |
GO:0071798 |
response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 |
0.1 |
GO:0018894 |
dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 |
0.9 |
GO:0006527 |
arginine catabolic process(GO:0006527) |
| 0.0 |
0.1 |
GO:0002752 |
leukocyte chemotaxis involved in inflammatory response(GO:0002232) cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 |
0.3 |
GO:1903237 |
negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 |
0.4 |
GO:0006627 |
protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 |
0.2 |
GO:0002606 |
positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 |
0.1 |
GO:0014876 |
response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.0 |
0.1 |
GO:2000974 |
auditory receptor cell fate determination(GO:0042668) negative regulation of mechanoreceptor differentiation(GO:0045632) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.0 |
0.1 |
GO:0015993 |
L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 |
0.1 |
GO:0033198 |
response to ATP(GO:0033198) |
| 0.0 |
0.2 |
GO:0002415 |
immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 |
0.1 |
GO:0035915 |
pore formation in membrane of other organism(GO:0035915) |
| 0.0 |
0.3 |
GO:1901908 |
diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 |
0.1 |
GO:1903697 |
negative regulation of microvillus assembly(GO:1903697) |
| 0.0 |
0.1 |
GO:0000415 |
negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 |
0.2 |
GO:0033277 |
abortive mitotic cell cycle(GO:0033277) |
| 0.0 |
0.1 |
GO:0010360 |
negative regulation of anion channel activity(GO:0010360) negative regulation of chloride transport(GO:2001226) |
| 0.0 |
0.3 |
GO:1990822 |
basic amino acid transmembrane transport(GO:1990822) |
| 0.0 |
0.9 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 |
0.2 |
GO:1904158 |
axonemal central apparatus assembly(GO:1904158) |
| 0.0 |
0.1 |
GO:0030327 |
prenylated protein catabolic process(GO:0030327) |
| 0.0 |
0.2 |
GO:0072658 |
maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 |
0.7 |
GO:0070986 |
left/right axis specification(GO:0070986) |
| 0.0 |
0.2 |
GO:1901624 |
negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.0 |
0.2 |
GO:2000503 |
positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 |
0.1 |
GO:0042495 |
detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 |
0.2 |
GO:0060935 |
transforming growth factor beta receptor complex assembly(GO:0007181) cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 0.0 |
0.2 |
GO:0015917 |
aminophospholipid transport(GO:0015917) |
| 0.0 |
0.0 |
GO:0030320 |
cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) cellular phosphate ion homeostasis(GO:0030643) phosphate ion homeostasis(GO:0055062) monovalent inorganic anion homeostasis(GO:0055083) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) divalent inorganic anion homeostasis(GO:0072505) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.0 |
0.1 |
GO:1902004 |
positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 |
0.3 |
GO:0001561 |
fatty acid alpha-oxidation(GO:0001561) |
| 0.0 |
0.1 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 |
0.0 |
GO:0042756 |
drinking behavior(GO:0042756) |
| 0.0 |
0.2 |
GO:0019918 |
peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 |
0.1 |
GO:0002337 |
B-1a B cell differentiation(GO:0002337) |
| 0.0 |
0.2 |
GO:0033387 |
putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 |
0.2 |
GO:0070560 |
protein secretion by platelet(GO:0070560) |
| 0.0 |
0.1 |
GO:0045925 |
positive regulation of female receptivity(GO:0045925) |
| 0.0 |
0.5 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.0 |
0.0 |
GO:0050965 |
detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 |
0.2 |
GO:0035407 |
histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 |
0.1 |
GO:0035604 |
fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 0.0 |
0.1 |
GO:0003064 |
regulation of heart rate by hormone(GO:0003064) |
| 0.0 |
0.6 |
GO:0002862 |
negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 |
0.1 |
GO:1903207 |
neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.0 |
0.1 |
GO:0034553 |
respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 |
0.1 |
GO:0051835 |
positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 |
0.3 |
GO:0035754 |
B cell chemotaxis(GO:0035754) |
| 0.0 |
0.1 |
GO:0071469 |
detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.0 |
0.1 |
GO:0009443 |
pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 |
0.4 |
GO:0009414 |
response to water deprivation(GO:0009414) |
| 0.0 |
0.6 |
GO:0003084 |
positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 |
0.5 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.0 |
0.1 |
GO:0032474 |
otolith morphogenesis(GO:0032474) |
| 0.0 |
0.3 |
GO:0086024 |
adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 |
0.1 |
GO:0090675 |
intermicrovillar adhesion(GO:0090675) |
| 0.0 |
0.3 |
GO:0098735 |
positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 |
0.3 |
GO:0032286 |
central nervous system myelin maintenance(GO:0032286) |
| 0.0 |
0.1 |
GO:0060084 |
synaptic transmission involved in micturition(GO:0060084) |
| 0.0 |
0.0 |
GO:0021527 |
spinal cord association neuron differentiation(GO:0021527) |
| 0.0 |
0.4 |
GO:0006600 |
creatine metabolic process(GO:0006600) |
| 0.0 |
0.2 |
GO:0038169 |
somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 |
0.1 |
GO:2001245 |
regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 |
0.3 |
GO:0072592 |
oxygen metabolic process(GO:0072592) |
| 0.0 |
0.6 |
GO:1901750 |
leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 |
0.1 |
GO:0001560 |
regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 |
0.5 |
GO:0038203 |
TORC2 signaling(GO:0038203) |
| 0.0 |
0.1 |
GO:1904031 |
positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 |
0.6 |
GO:0032988 |
ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 |
0.2 |
GO:0002032 |
desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 |
0.1 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.0 |
0.1 |
GO:1904647 |
response to rotenone(GO:1904647) |
| 0.0 |
0.1 |
GO:0007497 |
posterior midgut development(GO:0007497) |
| 0.0 |
0.1 |
GO:0043602 |
nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.0 |
0.7 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.0 |
0.1 |
GO:0015891 |
iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 |
0.3 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.0 |
0.0 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 0.0 |
0.1 |
GO:0046909 |
intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 |
0.1 |
GO:0043385 |
mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.0 |
0.3 |
GO:1903546 |
protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 |
0.2 |
GO:0097112 |
gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 |
0.0 |
GO:1990926 |
short-term synaptic potentiation(GO:1990926) |
| 0.0 |
0.1 |
GO:0032484 |
Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 |
0.1 |
GO:1990569 |
UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 |
0.1 |
GO:0007538 |
primary sex determination(GO:0007538) |
| 0.0 |
0.2 |
GO:0045653 |
negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 |
0.2 |
GO:0072602 |
interleukin-4 secretion(GO:0072602) |
| 0.0 |
0.2 |
GO:0072513 |
positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 |
0.1 |
GO:0043311 |
positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 |
0.1 |
GO:1903413 |
cellular response to bile acid(GO:1903413) |
| 0.0 |
0.3 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 |
0.5 |
GO:0046185 |
aldehyde catabolic process(GO:0046185) |
| 0.0 |
0.1 |
GO:2000661 |
positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 |
0.1 |
GO:0051958 |
methotrexate transport(GO:0051958) |
| 0.0 |
0.1 |
GO:1902361 |
mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 |
0.1 |
GO:0090156 |
cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 |
0.1 |
GO:0031296 |
B cell costimulation(GO:0031296) |
| 0.0 |
0.1 |
GO:0060988 |
lipid tube assembly(GO:0060988) |
| 0.0 |
0.1 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.0 |
0.1 |
GO:0042350 |
GDP-L-fucose biosynthetic process(GO:0042350) |
| 0.0 |
0.2 |
GO:0032380 |
regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 |
0.2 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 |
0.2 |
GO:0009450 |
acetate metabolic process(GO:0006083) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.0 |
0.2 |
GO:0003025 |
regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.0 |
0.6 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.0 |
0.2 |
GO:0055005 |
ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 |
0.1 |
GO:0035635 |
entry of bacterium into host cell(GO:0035635) |
| 0.0 |
0.3 |
GO:0015889 |
cobalamin transport(GO:0015889) |
| 0.0 |
0.1 |
GO:0043012 |
regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 |
0.1 |
GO:1904588 |
cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.0 |
0.6 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 |
0.3 |
GO:2000304 |
positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 |
0.1 |
GO:0048663 |
neuron fate commitment(GO:0048663) |
| 0.0 |
0.0 |
GO:0060775 |
mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.0 |
0.1 |
GO:0010165 |
response to X-ray(GO:0010165) |
| 0.0 |
0.2 |
GO:0061470 |
T follicular helper cell differentiation(GO:0061470) |
| 0.0 |
0.7 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
0.1 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 |
0.2 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 |
0.3 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.0 |
0.0 |
GO:0007206 |
phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 |
0.1 |
GO:0009415 |
response to water(GO:0009415) response to hydrostatic pressure(GO:0051599) |
| 0.0 |
3.8 |
GO:0042633 |
molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.0 |
0.0 |
GO:1904882 |
telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 |
0.1 |
GO:0048203 |
vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 |
0.1 |
GO:0072675 |
multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 |
0.0 |
GO:0002572 |
pro-T cell differentiation(GO:0002572) |
| 0.0 |
0.1 |
GO:1902723 |
negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 |
0.1 |
GO:0060010 |
Sertoli cell fate commitment(GO:0060010) |
| 0.0 |
0.3 |
GO:0070070 |
proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 |
0.1 |
GO:0060385 |
axonogenesis involved in innervation(GO:0060385) |
| 0.0 |
0.0 |
GO:2000194 |
regulation of female gonad development(GO:2000194) |
| 0.0 |
0.1 |
GO:0072233 |
thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 |
0.3 |
GO:0031848 |
protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 |
0.2 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.0 |
0.1 |
GO:0006041 |
glucosamine metabolic process(GO:0006041) |
| 0.0 |
0.4 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.0 |
0.2 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 |
0.1 |
GO:2000563 |
positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 |
0.1 |
GO:0034184 |
positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 |
0.4 |
GO:0042789 |
mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 |
0.0 |
GO:0002277 |
myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.0 |
0.1 |
GO:0071895 |
odontoblast differentiation(GO:0071895) |
| 0.0 |
0.4 |
GO:0051255 |
spindle midzone assembly(GO:0051255) |
| 0.0 |
0.1 |
GO:0019230 |
proprioception(GO:0019230) |
| 0.0 |
0.1 |
GO:0021514 |
ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 |
0.1 |
GO:0060373 |
regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 |
0.1 |
GO:0006097 |
glyoxylate cycle(GO:0006097) |
| 0.0 |
0.0 |
GO:2000389 |
regulation of neutrophil extravasation(GO:2000389) |
| 0.0 |
0.1 |
GO:0033615 |
mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 |
0.4 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 |
0.3 |
GO:0006782 |
protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 |
0.0 |
GO:0042853 |
L-alanine metabolic process(GO:0042851) L-alanine catabolic process(GO:0042853) |
| 0.0 |
0.0 |
GO:0043388 |
positive regulation of DNA binding(GO:0043388) |
| 0.0 |
0.5 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 |
0.1 |
GO:0048073 |
regulation of eye pigmentation(GO:0048073) |
| 0.0 |
0.1 |
GO:0036353 |
histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 |
0.1 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 |
0.2 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.0 |
0.1 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 |
0.1 |
GO:0060754 |
positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 |
0.1 |
GO:0008637 |
apoptotic mitochondrial changes(GO:0008637) |
| 0.0 |
0.1 |
GO:1901491 |
negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 |
0.1 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 |
0.0 |
GO:0050917 |
sensory perception of umami taste(GO:0050917) |
| 0.0 |
0.1 |
GO:0042769 |
DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 |
0.1 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.0 |
1.3 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 |
0.1 |
GO:0032416 |
negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.0 |
0.2 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 |
0.1 |
GO:0007023 |
post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 |
0.1 |
GO:0014016 |
neuroblast differentiation(GO:0014016) |
| 0.0 |
0.1 |
GO:0044240 |
multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 |
0.1 |
GO:0032096 |
negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 |
0.1 |
GO:1901569 |
leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 |
0.2 |
GO:1902510 |
regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.0 |
0.3 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.0 |
0.1 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 |
0.0 |
GO:0003186 |
tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) |
| 0.0 |
1.3 |
GO:0001895 |
retina homeostasis(GO:0001895) |
| 0.0 |
0.0 |
GO:0010818 |
T cell chemotaxis(GO:0010818) |
| 0.0 |
0.1 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.0 |
0.2 |
GO:2000427 |
positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 |
0.2 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 |
0.1 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 |
0.1 |
GO:2000676 |
positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 |
0.1 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 |
0.1 |
GO:0098989 |
NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 |
0.2 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 |
0.1 |
GO:0033563 |
dorsal/ventral axon guidance(GO:0033563) |
| 0.0 |
0.1 |
GO:0034311 |
diol metabolic process(GO:0034311) |
| 0.0 |
0.1 |
GO:0046952 |
ketone body catabolic process(GO:0046952) |
| 0.0 |
0.1 |
GO:1902031 |
regulation of NADP metabolic process(GO:1902031) |
| 0.0 |
0.1 |
GO:0006740 |
NADPH regeneration(GO:0006740) |
| 0.0 |
0.0 |
GO:1990086 |
lens fiber cell apoptotic process(GO:1990086) |
| 0.0 |
0.2 |
GO:0016188 |
synaptic vesicle maturation(GO:0016188) |
| 0.0 |
0.2 |
GO:0002315 |
marginal zone B cell differentiation(GO:0002315) |
| 0.0 |
0.1 |
GO:2000721 |
positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 |
0.1 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 |
0.1 |
GO:0048105 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.0 |
0.7 |
GO:0015991 |
ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 |
0.2 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.0 |
0.1 |
GO:1904383 |
response to sodium phosphate(GO:1904383) |
| 0.0 |
0.1 |
GO:0035523 |
protein K29-linked deubiquitination(GO:0035523) protein K6-linked deubiquitination(GO:0044313) |
| 0.0 |
0.0 |
GO:2000523 |
regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 |
0.1 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 |
0.0 |
GO:0030201 |
heparan sulfate proteoglycan metabolic process(GO:0030201) |
| 0.0 |
0.1 |
GO:2001170 |
negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 |
0.0 |
GO:0060338 |
regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 |
0.0 |
GO:0045216 |
cell-cell junction organization(GO:0045216) |
| 0.0 |
0.4 |
GO:0035988 |
chondrocyte proliferation(GO:0035988) |
| 0.0 |
0.2 |
GO:0010248 |
establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 |
0.2 |
GO:1903432 |
regulation of TORC1 signaling(GO:1903432) |
| 0.0 |
0.3 |
GO:0060716 |
labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 |
0.1 |
GO:0090481 |
pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 |
0.1 |
GO:0070383 |
DNA cytosine deamination(GO:0070383) |
| 0.0 |
0.2 |
GO:0090360 |
platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 |
0.1 |
GO:0070278 |
extracellular matrix constituent secretion(GO:0070278) |
| 0.0 |
0.0 |
GO:0006121 |
mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 |
0.0 |
GO:0035437 |
maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 |
0.1 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 |
0.1 |
GO:0060633 |
negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 0.0 |
0.1 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 |
0.2 |
GO:0038063 |
collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 |
0.2 |
GO:0033008 |
positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 |
0.0 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.0 |
0.4 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.0 |
0.1 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.0 |
0.2 |
GO:0070493 |
thrombin receptor signaling pathway(GO:0070493) |
| 0.0 |
0.1 |
GO:0006069 |
ethanol oxidation(GO:0006069) |
| 0.0 |
0.1 |
GO:0008582 |
regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 |
0.1 |
GO:0032571 |
response to vitamin K(GO:0032571) |
| 0.0 |
0.3 |
GO:2000651 |
positive regulation of sodium ion transmembrane transport(GO:1902307) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 |
0.1 |
GO:0071847 |
TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 |
0.1 |
GO:0035608 |
protein deglutamylation(GO:0035608) |
| 0.0 |
0.1 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 |
0.4 |
GO:0051156 |
glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 |
0.0 |
GO:0019682 |
glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 |
0.2 |
GO:0001514 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 |
0.1 |
GO:0045872 |
regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 |
0.2 |
GO:0048003 |
antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 |
0.1 |
GO:2000857 |
positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 |
0.1 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 |
0.0 |
GO:0007266 |
Rho protein signal transduction(GO:0007266) |
| 0.0 |
0.1 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 |
0.2 |
GO:0045329 |
carnitine biosynthetic process(GO:0045329) |
| 0.0 |
0.1 |
GO:0043324 |
eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 |
0.0 |
GO:0034182 |
regulation of maintenance of sister chromatid cohesion(GO:0034091) regulation of maintenance of mitotic sister chromatid cohesion(GO:0034182) |
| 0.0 |
0.0 |
GO:0016095 |
polyprenol catabolic process(GO:0016095) |
| 0.0 |
0.0 |
GO:1901052 |
sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.0 |
0.1 |
GO:1902715 |
positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 |
0.5 |
GO:0003341 |
cilium movement(GO:0003341) |
| 0.0 |
0.1 |
GO:0031580 |
membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 |
0.1 |
GO:0001955 |
blood vessel maturation(GO:0001955) |
| 0.0 |
0.7 |
GO:1904837 |
beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 |
0.1 |
GO:0019566 |
arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 |
0.1 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.0 |
0.1 |
GO:0036289 |
peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 |
0.1 |
GO:0018094 |
protein polyglycylation(GO:0018094) |
| 0.0 |
0.1 |
GO:0032516 |
positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 |
0.0 |
GO:0034427 |
nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 |
0.1 |
GO:2000418 |
positive regulation of eosinophil migration(GO:2000418) |
| 0.0 |
0.1 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.0 |
0.1 |
GO:0090521 |
glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 |
0.1 |
GO:0035553 |
oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 |
0.1 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 |
0.0 |
GO:0043932 |
ossification involved in bone remodeling(GO:0043932) |
| 0.0 |
0.0 |
GO:0070676 |
intralumenal vesicle formation(GO:0070676) |
| 0.0 |
0.1 |
GO:0016559 |
peroxisome fission(GO:0016559) |
| 0.0 |
0.1 |
GO:0097211 |
response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 |
0.1 |
GO:0071400 |
cellular response to oleic acid(GO:0071400) |
| 0.0 |
0.1 |
GO:0051552 |
flavone metabolic process(GO:0051552) |
| 0.0 |
0.0 |
GO:0040009 |
regulation of growth rate(GO:0040009) |
| 0.0 |
0.0 |
GO:0061304 |
retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 |
0.0 |
GO:0009624 |
response to nematode(GO:0009624) |
| 0.0 |
0.3 |
GO:0043488 |
regulation of mRNA stability(GO:0043488) |
| 0.0 |
0.0 |
GO:1901731 |
lymphatic endothelial cell fate commitment(GO:0060838) positive regulation of platelet aggregation(GO:1901731) regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.0 |
0.2 |
GO:0035372 |
protein localization to microtubule(GO:0035372) |
| 0.0 |
0.2 |
GO:0002031 |
G-protein coupled receptor internalization(GO:0002031) |
| 0.0 |
0.0 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.0 |
0.2 |
GO:0070830 |
bicellular tight junction assembly(GO:0070830) |
| 0.0 |
0.2 |
GO:0035826 |
rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 |
0.1 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
0.3 |
GO:0006895 |
Golgi to endosome transport(GO:0006895) |
| 0.0 |
0.1 |
GO:0048278 |
vesicle docking(GO:0048278) |
| 0.0 |
0.1 |
GO:0051974 |
negative regulation of telomerase activity(GO:0051974) |
| 0.0 |
0.7 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.0 |
0.0 |
GO:0015781 |
pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 |
0.1 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 |
0.0 |
GO:0070373 |
negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 |
0.3 |
GO:0097242 |
beta-amyloid clearance(GO:0097242) |
| 0.0 |
0.1 |
GO:0060050 |
positive regulation of protein glycosylation(GO:0060050) |
| 0.0 |
0.0 |
GO:1903966 |
monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 |
0.1 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.0 |
0.4 |
GO:0006198 |
cAMP catabolic process(GO:0006198) |
| 0.0 |
0.0 |
GO:0002693 |
positive regulation of cellular extravasation(GO:0002693) |
| 0.0 |
0.2 |
GO:0010804 |
negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 |
0.1 |
GO:0048194 |
Golgi vesicle budding(GO:0048194) |
| 0.0 |
0.1 |
GO:0015842 |
aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 |
0.1 |
GO:0039536 |
negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 |
0.1 |
GO:0006307 |
DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 |
0.4 |
GO:0032007 |
negative regulation of TOR signaling(GO:0032007) |
| 0.0 |
0.1 |
GO:0032621 |
interleukin-18 production(GO:0032621) |
| 0.0 |
0.1 |
GO:1900113 |
negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 |
0.0 |
GO:0045776 |
negative regulation of blood pressure(GO:0045776) |
| 0.0 |
0.0 |
GO:0072709 |
cellular response to sorbitol(GO:0072709) |
| 0.0 |
0.1 |
GO:0048680 |
positive regulation of axon regeneration(GO:0048680) |
| 0.0 |
0.3 |
GO:0071801 |
regulation of podosome assembly(GO:0071801) |
| 0.0 |
0.1 |
GO:0046485 |
ether lipid metabolic process(GO:0046485) |
| 0.0 |
0.0 |
GO:0032755 |
positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 |
0.1 |
GO:0014005 |
microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 |
0.1 |
GO:0070127 |
tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 |
0.0 |
GO:1901300 |
positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 |
0.0 |
GO:0006642 |
triglyceride mobilization(GO:0006642) |
| 0.0 |
0.1 |
GO:0044341 |
sodium-dependent phosphate transport(GO:0044341) |
| 0.0 |
0.1 |
GO:0032025 |
response to cobalt ion(GO:0032025) |
| 0.0 |
0.1 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.0 |
0.1 |
GO:0060071 |
Wnt signaling pathway, planar cell polarity pathway(GO:0060071) regulation of establishment of planar polarity(GO:0090175) |
| 0.0 |
0.0 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.0 |
0.1 |
GO:0045779 |
negative regulation of bone resorption(GO:0045779) |
| 0.0 |
0.3 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 |
0.4 |
GO:0045022 |
early endosome to late endosome transport(GO:0045022) |
| 0.0 |
0.0 |
GO:0031507 |
heterochromatin assembly(GO:0031507) |
| 0.0 |
0.3 |
GO:0090140 |
regulation of mitochondrial fission(GO:0090140) |
| 0.0 |
0.1 |
GO:0019626 |
short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 |
0.1 |
GO:0014816 |
skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 |
0.2 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 |
0.0 |
GO:0097267 |
omega-hydroxylase P450 pathway(GO:0097267) |
| 0.0 |
0.2 |
GO:0042447 |
hormone catabolic process(GO:0042447) |
| 0.0 |
0.2 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.1 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 |
0.1 |
GO:0006772 |
thiamine metabolic process(GO:0006772) |
| 0.0 |
0.3 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.0 |
0.2 |
GO:0032091 |
negative regulation of protein binding(GO:0032091) |
| 0.0 |
0.1 |
GO:0060412 |
ventricular septum morphogenesis(GO:0060412) |