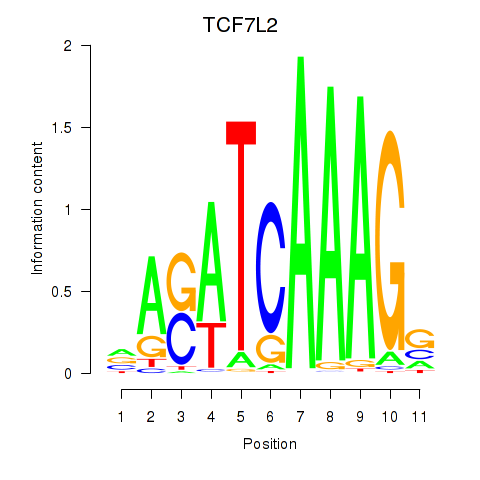
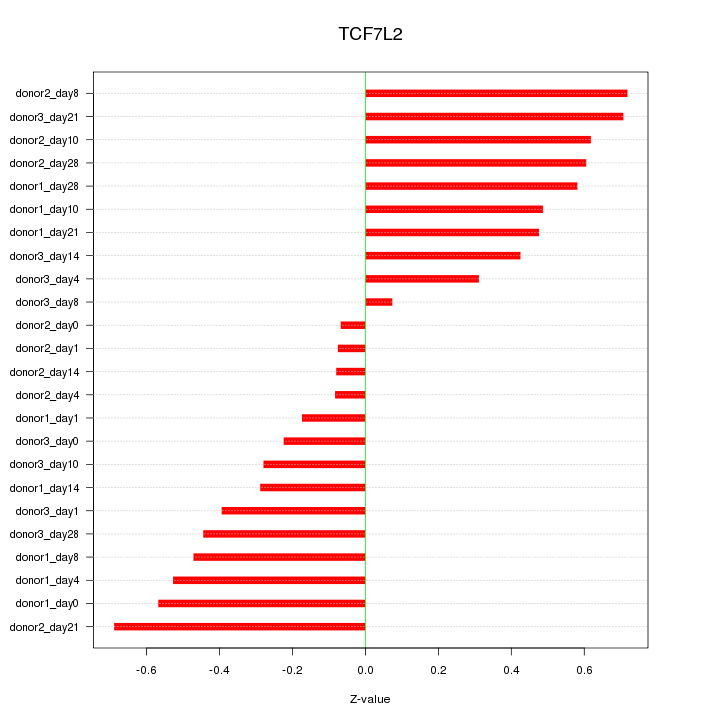
| 1.1 |
3.3 |
GO:0061181 |
regulation of chondrocyte development(GO:0061181) |
| 0.4 |
1.1 |
GO:1903568 |
negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.2 |
0.7 |
GO:1901202 |
negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.1 |
0.3 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 |
0.3 |
GO:0072658 |
maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 |
0.3 |
GO:0035426 |
extracellular matrix-cell signaling(GO:0035426) |
| 0.1 |
0.2 |
GO:0060086 |
circadian temperature homeostasis(GO:0060086) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.1 |
0.2 |
GO:1904338 |
regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 |
0.2 |
GO:1904059 |
regulation of locomotor rhythm(GO:1904059) |
| 0.0 |
0.2 |
GO:0018032 |
protein amidation(GO:0018032) |
| 0.0 |
0.2 |
GO:2000729 |
positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 |
0.2 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 |
0.1 |
GO:0003218 |
cardiac left ventricle formation(GO:0003218) |
| 0.0 |
0.2 |
GO:0002314 |
germinal center B cell differentiation(GO:0002314) |
| 0.0 |
0.2 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 |
0.2 |
GO:0044336 |
canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 |
0.3 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 |
0.2 |
GO:0010961 |
cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 |
0.2 |
GO:0019626 |
short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 |
0.2 |
GO:2000312 |
regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 |
0.1 |
GO:0042489 |
negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.0 |
0.1 |
GO:0015729 |
thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.0 |
0.4 |
GO:0045078 |
positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 |
0.2 |
GO:0001865 |
NK T cell differentiation(GO:0001865) |
| 0.0 |
0.2 |
GO:0055059 |
asymmetric neuroblast division(GO:0055059) |
| 0.0 |
0.1 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 |
0.5 |
GO:0002517 |
T cell tolerance induction(GO:0002517) |
| 0.0 |
0.4 |
GO:0035372 |
protein localization to microtubule(GO:0035372) |
| 0.0 |
0.6 |
GO:0072189 |
ureter development(GO:0072189) |
| 0.0 |
0.3 |
GO:0006600 |
creatine metabolic process(GO:0006600) |
| 0.0 |
0.1 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 |
0.1 |
GO:0071279 |
cellular response to cobalt ion(GO:0071279) |
| 0.0 |
0.4 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) |
| 0.0 |
0.1 |
GO:0006041 |
glucosamine metabolic process(GO:0006041) |
| 0.0 |
0.8 |
GO:0021952 |
central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 |
0.1 |
GO:0060244 |
negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 |
0.0 |
GO:0003192 |
mitral valve formation(GO:0003192) |
| 0.0 |
0.1 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) |
| 0.0 |
0.0 |
GO:0003129 |
heart induction(GO:0003129) negative regulation of mesodermal cell fate specification(GO:0042662) regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 |
1.9 |
GO:0001942 |
hair follicle development(GO:0001942) |
| 0.0 |
0.1 |
GO:0021615 |
glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 |
0.3 |
GO:0003334 |
keratinocyte development(GO:0003334) |
| 0.0 |
0.7 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.4 |
GO:0035338 |
long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 |
0.1 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 |
0.3 |
GO:0034389 |
lipid particle organization(GO:0034389) |
| 0.0 |
0.1 |
GO:0014807 |
regulation of somitogenesis(GO:0014807) |
| 0.0 |
0.0 |
GO:0032661 |
regulation of interleukin-18 production(GO:0032661) |
| 0.0 |
0.1 |
GO:0032511 |
late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |