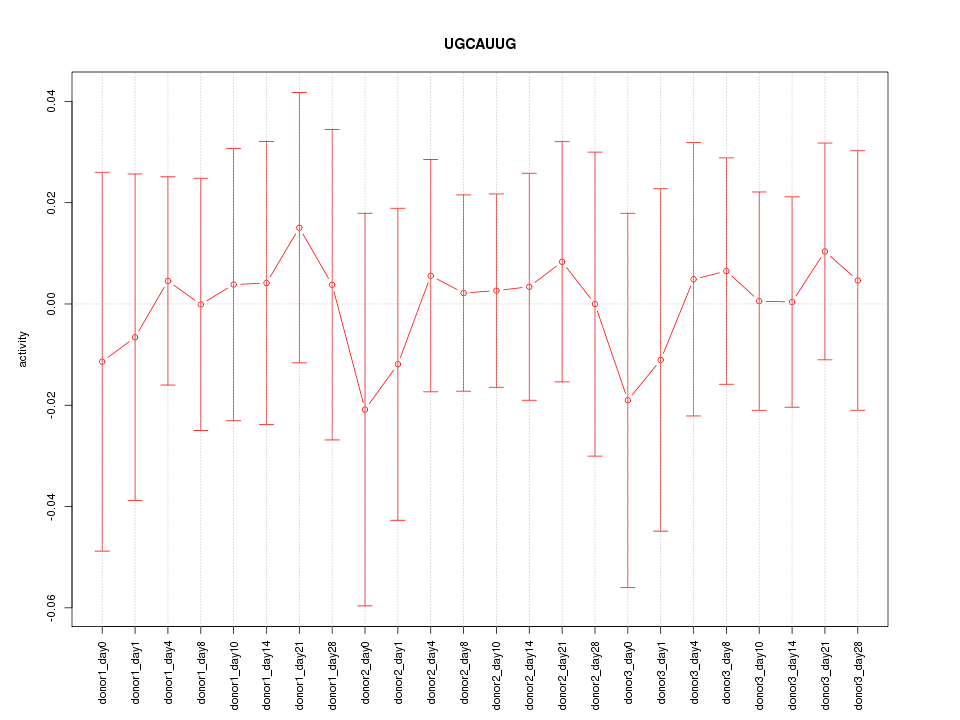
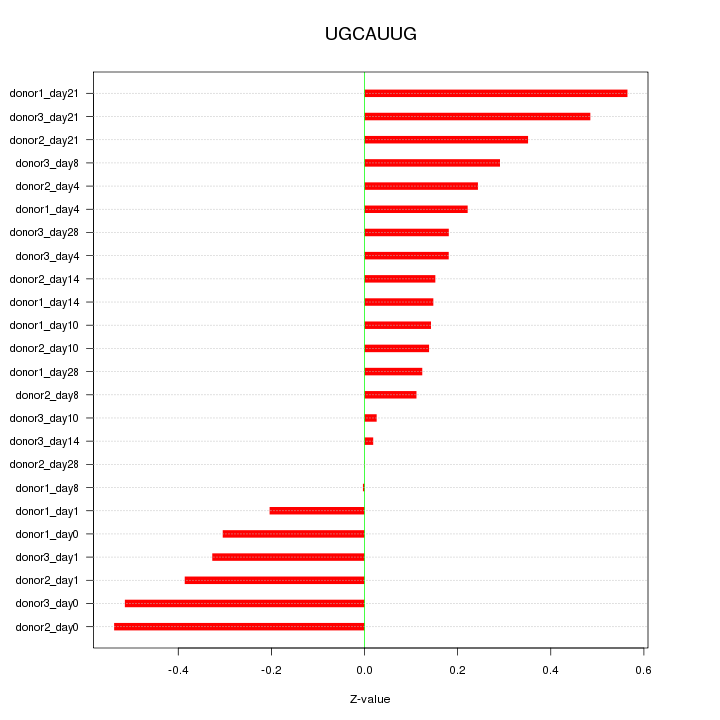
| 0.1 |
0.2 |
GO:0002071 |
glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.1 |
0.3 |
GO:0000255 |
allantoin metabolic process(GO:0000255) |
| 0.1 |
0.2 |
GO:0035470 |
positive regulation of vascular wound healing(GO:0035470) regulation of lactation(GO:1903487) |
| 0.0 |
0.1 |
GO:0097156 |
fasciculation of motor neuron axon(GO:0097156) |
| 0.0 |
0.1 |
GO:1900169 |
regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 |
0.5 |
GO:0060628 |
regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 |
0.1 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 |
0.1 |
GO:0045925 |
positive regulation of female receptivity(GO:0045925) |
| 0.0 |
0.1 |
GO:0071630 |
nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 |
0.1 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 |
0.2 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 |
0.2 |
GO:0032000 |
positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 |
0.0 |
GO:0035494 |
SNARE complex disassembly(GO:0035494) |
| 0.0 |
0.1 |
GO:0001555 |
oocyte growth(GO:0001555) |
| 0.0 |
0.1 |
GO:0072658 |
maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 |
2.9 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.1 |
GO:0045964 |
positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 |
0.1 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.0 |
0.1 |
GO:0008063 |
Toll signaling pathway(GO:0008063) |
| 0.0 |
0.3 |
GO:2001275 |
positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 |
0.1 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 |
0.1 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.0 |
0.0 |
GO:0099543 |
retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 |
0.0 |
GO:0002101 |
tRNA wobble cytosine modification(GO:0002101) oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 |
0.1 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.0 |
0.0 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 |
0.1 |
GO:0060235 |
lens induction in camera-type eye(GO:0060235) |
| 0.0 |
0.0 |
GO:0006679 |
glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 |
0.2 |
GO:0015866 |
ADP transport(GO:0015866) |
| 0.0 |
0.0 |
GO:1904016 |
response to Thyroglobulin triiodothyronine(GO:1904016) |
| 0.0 |
0.2 |
GO:0072189 |
ureter development(GO:0072189) |