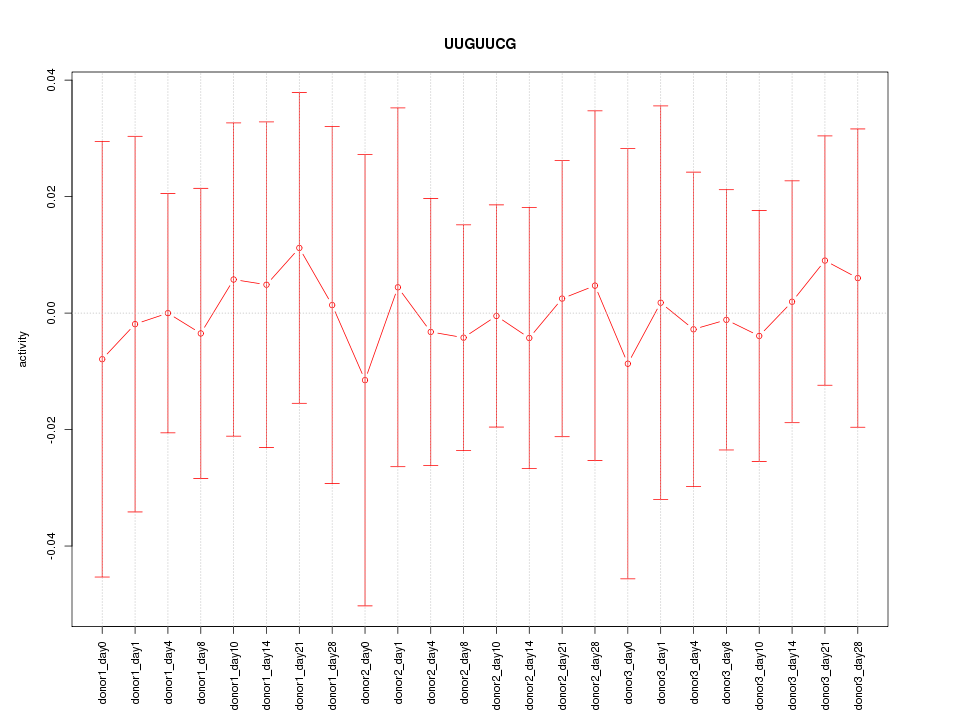
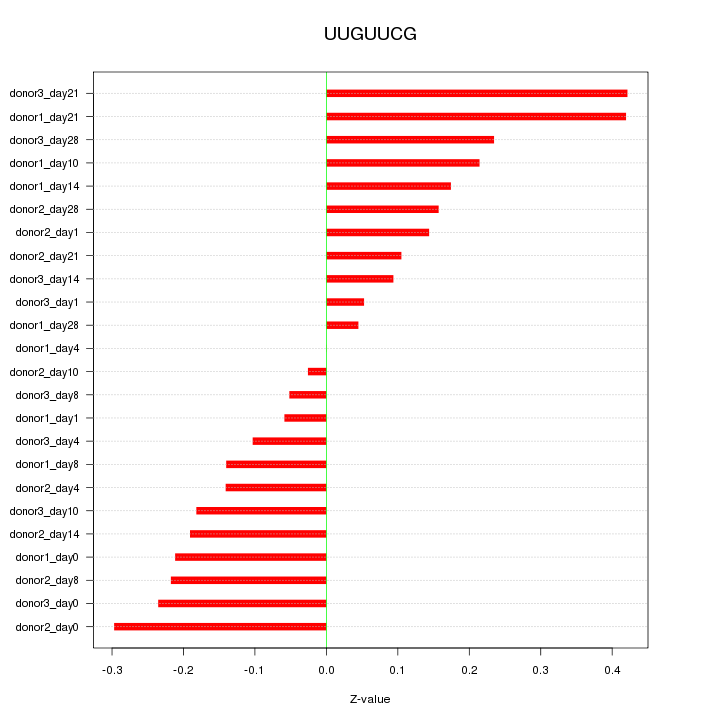
|
chr14_+_74486043
|
0.203
|
ENST00000464394.1
ENST00000394009.3
|
CCDC176
|
coiled-coil domain containing 176
|
|
chr3_-_114790179
|
0.162
|
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr17_+_14204389
|
0.128
|
ENST00000360954.2
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr19_-_14316980
|
0.125
|
ENST00000361434.3
ENST00000340736.6
|
LPHN1
|
latrophilin 1
|
|
chr11_-_74178582
|
0.125
|
ENST00000531854.1
ENST00000526855.1
ENST00000529425.1
ENST00000310128.4
ENST00000532569.1
|
KCNE3
|
potassium voltage-gated channel, Isk-related family, member 3
|
|
chrX_+_95939711
|
0.107
|
ENST00000373049.4
ENST00000324765.8
|
DIAPH2
|
diaphanous-related formin 2
|
|
chr6_+_159084188
|
0.094
|
ENST00000367081.3
|
SYTL3
|
synaptotagmin-like 3
|
|
chr2_-_172750733
|
0.093
|
ENST00000392592.4
ENST00000422440.2
|
SLC25A12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12
|
|
chr18_-_53255766
|
0.093
|
ENST00000566286.1
ENST00000564999.1
ENST00000566279.1
ENST00000354452.3
ENST00000356073.4
|
TCF4
|
transcription factor 4
|
|
chr18_+_13218769
|
0.091
|
ENST00000399848.3
ENST00000361205.4
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4
|
|
chr1_+_61547894
|
0.090
|
ENST00000403491.3
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_180601139
|
0.089
|
ENST00000367590.4
ENST00000367589.3
|
XPR1
|
xenotropic and polytropic retrovirus receptor 1
|
|
chr13_-_31736027
|
0.086
|
ENST00000380406.5
ENST00000320027.5
ENST00000380405.4
|
HSPH1
|
heat shock 105kDa/110kDa protein 1
|
|
chr2_+_179345173
|
0.084
|
ENST00000234453.5
|
PLEKHA3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3
|
|
chr1_-_213189108
|
0.080
|
ENST00000535388.1
|
ANGEL2
|
angel homolog 2 (Drosophila)
|
|
chr11_-_86666427
|
0.072
|
ENST00000531380.1
|
FZD4
|
frizzled family receptor 4
|
|
chr16_+_86544113
|
0.072
|
ENST00000262426.4
|
FOXF1
|
forkhead box F1
|
|
chrX_-_100604184
|
0.071
|
ENST00000372902.3
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast)
|
|
chr14_+_57735614
|
0.070
|
ENST00000261558.3
|
AP5M1
|
adaptor-related protein complex 5, mu 1 subunit
|
|
chr7_+_17338239
|
0.067
|
ENST00000242057.4
|
AHR
|
aryl hydrocarbon receptor
|
|
chr9_+_26956371
|
0.067
|
ENST00000380062.5
ENST00000518614.1
|
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas)
|
|
chr6_+_149068464
|
0.065
|
ENST00000367463.4
|
UST
|
uronyl-2-sulfotransferase
|
|
chr19_-_7293942
|
0.065
|
ENST00000341500.5
ENST00000302850.5
|
INSR
|
insulin receptor
|
|
chr4_+_72204755
|
0.063
|
ENST00000512686.1
ENST00000340595.3
|
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4
|
|
chr6_+_4021554
|
0.062
|
ENST00000337659.6
|
PRPF4B
|
pre-mRNA processing factor 4B
|
|
chr17_-_7832753
|
0.060
|
ENST00000303790.2
|
KCNAB3
|
potassium voltage-gated channel, shaker-related subfamily, beta member 3
|
|
chr9_-_135230336
|
0.060
|
ENST00000224140.5
ENST00000372169.2
ENST00000393220.1
|
SETX
|
senataxin
|
|
chr13_-_95364389
|
0.059
|
ENST00000376945.2
|
SOX21
|
SRY (sex determining region Y)-box 21
|
|
chr3_+_197687071
|
0.057
|
ENST00000482695.1
ENST00000330198.4
ENST00000419117.1
ENST00000420910.2
ENST00000332636.5
|
LMLN
|
leishmanolysin-like (metallopeptidase M8 family)
|
|
chr1_-_57045228
|
0.057
|
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr1_-_225840747
|
0.053
|
ENST00000366843.2
ENST00000366844.3
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr14_-_70883708
|
0.051
|
ENST00000256366.4
|
SYNJ2BP
|
synaptojanin 2 binding protein
|
|
chr5_+_56111361
|
0.050
|
ENST00000399503.3
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase
|
|
chr10_-_52383644
|
0.046
|
ENST00000361781.2
|
SGMS1
|
sphingomyelin synthase 1
|
|
chr7_-_130080977
|
0.046
|
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa
|
|
chr12_+_56401268
|
0.045
|
ENST00000262032.5
|
IKZF4
|
IKAROS family zinc finger 4 (Eos)
|
|
chr5_+_145583156
|
0.045
|
ENST00000265271.5
|
RBM27
|
RNA binding motif protein 27
|
|
chr7_+_75831181
|
0.045
|
ENST00000388802.4
ENST00000326382.8
|
SRRM3
|
serine/arginine repetitive matrix 3
|
|
chr16_+_77822427
|
0.045
|
ENST00000302536.2
|
VAT1L
|
vesicle amine transport 1-like
|
|
chr22_+_29168652
|
0.045
|
ENST00000249064.4
ENST00000444523.1
ENST00000448492.2
ENST00000421503.2
|
CCDC117
|
coiled-coil domain containing 117
|
|
chr5_+_149109825
|
0.045
|
ENST00000360453.4
ENST00000394320.3
ENST00000309241.5
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr16_+_2587998
|
0.044
|
ENST00000441549.3
ENST00000268673.7
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr10_-_89623194
|
0.044
|
ENST00000445946.3
|
KLLN
|
killin, p53-regulated DNA replication inhibitor
|
|
chrX_+_69664706
|
0.043
|
ENST00000194900.4
ENST00000374360.3
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr14_+_55493920
|
0.043
|
ENST00000395472.2
ENST00000555846.1
|
SOCS4
|
suppressor of cytokine signaling 4
|
|
chr20_+_43514315
|
0.042
|
ENST00000353703.4
|
YWHAB
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta
|
|
chr14_-_96830207
|
0.041
|
ENST00000359933.4
|
ATG2B
|
autophagy related 2B
|
|
chr17_-_27621125
|
0.041
|
ENST00000579665.1
ENST00000225388.4
|
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chr11_+_34127142
|
0.040
|
ENST00000257829.3
ENST00000531159.2
|
NAT10
|
N-acetyltransferase 10 (GCN5-related)
|
|
chr13_+_73632897
|
0.037
|
ENST00000377687.4
|
KLF5
|
Kruppel-like factor 5 (intestinal)
|
|
chr1_-_10856694
|
0.037
|
ENST00000377022.3
ENST00000344008.5
|
CASZ1
|
castor zinc finger 1
|
|
chr19_+_47421933
|
0.036
|
ENST00000404338.3
|
ARHGAP35
|
Rho GTPase activating protein 35
|
|
chr19_-_6279932
|
0.036
|
ENST00000252674.7
|
MLLT1
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1
|
|
chr17_+_55162453
|
0.034
|
ENST00000575322.1
ENST00000337714.3
ENST00000314126.3
|
AKAP1
|
A kinase (PRKA) anchor protein 1
|
|
chr2_+_133174147
|
0.033
|
ENST00000329321.3
|
GPR39
|
G protein-coupled receptor 39
|
|
chr21_-_38362497
|
0.032
|
ENST00000427746.1
ENST00000336648.4
|
HLCS
|
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase)
|
|
chr12_+_103351444
|
0.032
|
ENST00000266744.3
|
ASCL1
|
achaete-scute family bHLH transcription factor 1
|
|
chr9_-_127533519
|
0.031
|
ENST00000487099.2
ENST00000344523.4
ENST00000373584.3
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1
|
|
chr8_-_65711310
|
0.030
|
ENST00000310193.3
|
CYP7B1
|
cytochrome P450, family 7, subfamily B, polypeptide 1
|
|
chr7_-_27183263
|
0.030
|
ENST00000222726.3
|
HOXA5
|
homeobox A5
|
|
chr16_-_12897642
|
0.030
|
ENST00000433677.2
ENST00000261660.4
ENST00000381774.4
|
CPPED1
|
calcineurin-like phosphoesterase domain containing 1
|
|
chr17_+_28804380
|
0.030
|
ENST00000225724.5
ENST00000451249.2
ENST00000467337.2
ENST00000581721.1
ENST00000414833.2
|
GOSR1
|
golgi SNAP receptor complex member 1
|
|
chr4_-_83295103
|
0.029
|
ENST00000313899.7
ENST00000352301.4
ENST00000509107.1
ENST00000353341.4
ENST00000541060.1
|
HNRNPD
|
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa)
|
|
chr18_+_18943554
|
0.029
|
ENST00000580732.2
|
GREB1L
|
growth regulation by estrogen in breast cancer-like
|
|
chr2_-_178937478
|
0.029
|
ENST00000286063.6
|
PDE11A
|
phosphodiesterase 11A
|
|
chr15_-_72410109
|
0.028
|
ENST00000564571.1
|
MYO9A
|
myosin IXA
|
|
chr1_-_182573514
|
0.027
|
ENST00000367558.5
|
RGS16
|
regulator of G-protein signaling 16
|
|
chr16_+_69458428
|
0.027
|
ENST00000512062.1
ENST00000307892.8
|
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane)
|
|
chr4_+_113152881
|
0.027
|
ENST00000274000.5
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein
|
|
chr1_+_62902308
|
0.026
|
ENST00000339950.4
|
USP1
|
ubiquitin specific peptidase 1
|
|
chr17_-_3749515
|
0.026
|
ENST00000158149.3
ENST00000389005.4
|
C17orf85
|
chromosome 17 open reading frame 85
|
|
chr9_+_115983808
|
0.026
|
ENST00000374210.6
ENST00000374212.4
|
SLC31A1
|
solute carrier family 31 (copper transporter), member 1
|
|
chr1_-_21377447
|
0.025
|
ENST00000374937.3
ENST00000264211.8
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr2_+_242641442
|
0.025
|
ENST00000313552.6
ENST00000406941.1
|
ING5
|
inhibitor of growth family, member 5
|
|
chr4_-_40631859
|
0.024
|
ENST00000295971.7
ENST00000319592.4
|
RBM47
|
RNA binding motif protein 47
|
|
chr1_+_2985760
|
0.024
|
ENST00000378391.2
ENST00000514189.1
ENST00000270722.5
|
PRDM16
|
PR domain containing 16
|
|
chr7_-_105517021
|
0.023
|
ENST00000318724.4
ENST00000419735.3
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr3_-_135914615
|
0.023
|
ENST00000309993.2
|
MSL2
|
male-specific lethal 2 homolog (Drosophila)
|
|
chr10_+_45455207
|
0.022
|
ENST00000334940.6
ENST00000374417.2
ENST00000340258.5
ENST00000427758.1
|
RASSF4
|
Ras association (RalGDS/AF-6) domain family member 4
|
|
chr9_-_135819987
|
0.022
|
ENST00000298552.3
ENST00000403810.1
|
TSC1
|
tuberous sclerosis 1
|
|
chr13_+_51483814
|
0.022
|
ENST00000336617.3
ENST00000422660.1
|
RNASEH2B
|
ribonuclease H2, subunit B
|
|
chr16_+_56965960
|
0.022
|
ENST00000439977.2
ENST00000344114.4
ENST00000300302.5
ENST00000379792.2
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr17_-_60142609
|
0.021
|
ENST00000397786.2
|
MED13
|
mediator complex subunit 13
|
|
chr7_+_139026057
|
0.021
|
ENST00000541515.3
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae)
|
|
chr8_+_61591337
|
0.021
|
ENST00000423902.2
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr12_-_51477333
|
0.020
|
ENST00000228515.1
ENST00000548206.1
ENST00000546935.1
ENST00000548981.1
|
CSRNP2
|
cysteine-serine-rich nuclear protein 2
|
|
chr2_+_155554797
|
0.020
|
ENST00000295101.2
|
KCNJ3
|
potassium inwardly-rectifying channel, subfamily J, member 3
|
|
chr2_-_74405929
|
0.020
|
ENST00000396049.4
|
MOB1A
|
MOB kinase activator 1A
|
|
chr6_-_91006461
|
0.020
|
ENST00000257749.4
ENST00000343122.3
ENST00000406998.2
ENST00000453877.1
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr9_+_4679555
|
0.019
|
ENST00000381858.1
ENST00000381854.3
|
CDC37L1
|
cell division cycle 37-like 1
|
|
chr14_-_68283291
|
0.018
|
ENST00000555452.1
ENST00000347230.4
|
ZFYVE26
|
zinc finger, FYVE domain containing 26
|
|
chr10_-_126849068
|
0.017
|
ENST00000494626.2
ENST00000337195.5
|
CTBP2
|
C-terminal binding protein 2
|
|
chr3_-_151176497
|
0.016
|
ENST00000282466.3
|
IGSF10
|
immunoglobulin superfamily, member 10
|
|
chr8_-_38386175
|
0.015
|
ENST00000437935.2
ENST00000358138.1
|
C8orf86
|
chromosome 8 open reading frame 86
|
|
chr5_-_56247935
|
0.015
|
ENST00000381199.3
ENST00000381226.3
ENST00000381213.3
|
MIER3
|
mesoderm induction early response 1, family member 3
|
|
chr2_+_46926048
|
0.015
|
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr18_+_32558208
|
0.015
|
ENST00000436190.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr17_-_1465924
|
0.015
|
ENST00000573231.1
ENST00000576722.1
ENST00000576761.1
ENST00000576010.2
ENST00000313486.7
ENST00000539476.1
|
PITPNA
|
phosphatidylinositol transfer protein, alpha
|
|
chr9_-_16870704
|
0.015
|
ENST00000380672.4
ENST00000380667.2
ENST00000380666.2
ENST00000486514.1
|
BNC2
|
basonuclin 2
|
|
chr1_-_205601064
|
0.015
|
ENST00000357992.4
ENST00000289703.4
|
ELK4
|
ELK4, ETS-domain protein (SRF accessory protein 1)
|
|
chr9_-_21335356
|
0.014
|
ENST00000359039.4
|
KLHL9
|
kelch-like family member 9
|
|
chr1_-_67896095
|
0.014
|
ENST00000370994.4
|
SERBP1
|
SERPINE1 mRNA binding protein 1
|
|
chr12_-_51420128
|
0.014
|
ENST00000262051.7
ENST00000547732.1
ENST00000262052.5
ENST00000546488.1
ENST00000550714.1
ENST00000548193.1
ENST00000547579.1
ENST00000546743.1
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2
|
|
chr3_+_137483579
|
0.014
|
ENST00000306087.1
|
SOX14
|
SRY (sex determining region Y)-box 14
|
|
chr8_-_101965146
|
0.014
|
ENST00000395957.2
ENST00000395948.2
ENST00000457309.1
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta
|
|
chr12_-_14133053
|
0.014
|
ENST00000609686.1
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B
|
|
chr4_-_141677267
|
0.013
|
ENST00000442267.2
|
TBC1D9
|
TBC1 domain family, member 9 (with GRAM domain)
|
|
chr1_-_152332480
|
0.013
|
ENST00000388718.5
|
FLG2
|
filaggrin family member 2
|
|
chr17_+_77751931
|
0.013
|
ENST00000310942.4
ENST00000269399.5
|
CBX2
|
chromobox homolog 2
|
|
chr8_+_125551338
|
0.013
|
ENST00000276689.3
ENST00000518008.1
ENST00000522532.1
ENST00000517367.1
|
NDUFB9
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9, 22kDa
|
|
chr11_+_68080077
|
0.013
|
ENST00000294304.7
|
LRP5
|
low density lipoprotein receptor-related protein 5
|
|
chr17_+_27369918
|
0.012
|
ENST00000323372.4
|
PIPOX
|
pipecolic acid oxidase
|
|
chr6_-_16761678
|
0.012
|
ENST00000244769.4
ENST00000436367.1
|
ATXN1
|
ataxin 1
|
|
chr5_-_59189545
|
0.011
|
ENST00000340635.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr8_-_116681221
|
0.011
|
ENST00000395715.3
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr13_-_72441315
|
0.011
|
ENST00000305425.4
ENST00000313174.7
ENST00000354591.4
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr7_+_92158083
|
0.011
|
ENST00000265732.5
ENST00000481551.1
ENST00000496410.1
|
RBM48
|
RNA binding motif protein 48
|
|
chr5_+_138629417
|
0.010
|
ENST00000510056.1
ENST00000511249.1
ENST00000503811.1
ENST00000511378.1
|
MATR3
|
matrin 3
|
|
chr15_+_57210818
|
0.010
|
ENST00000438423.2
ENST00000267811.5
ENST00000452095.2
ENST00000559609.1
ENST00000333725.5
|
TCF12
|
transcription factor 12
|
|
chrX_-_49056635
|
0.009
|
ENST00000472598.1
ENST00000538567.1
ENST00000479808.1
ENST00000263233.4
|
SYP
|
synaptophysin
|
|
chr1_+_110009215
|
0.009
|
ENST00000369872.3
|
SYPL2
|
synaptophysin-like 2
|
|
chr15_+_68346501
|
0.009
|
ENST00000249636.6
|
PIAS1
|
protein inhibitor of activated STAT, 1
|
|
chr4_+_184020398
|
0.008
|
ENST00000403733.3
ENST00000378925.3
|
WWC2
|
WW and C2 domain containing 2
|
|
chr8_+_106330920
|
0.007
|
ENST00000407775.2
|
ZFPM2
|
zinc finger protein, FOG family member 2
|
|
chr8_-_117768023
|
0.007
|
ENST00000518949.1
ENST00000522453.1
ENST00000521861.1
ENST00000518995.1
|
EIF3H
|
eukaryotic translation initiation factor 3, subunit H
|
|
chr15_-_25684110
|
0.007
|
ENST00000232165.3
|
UBE3A
|
ubiquitin protein ligase E3A
|
|
chr19_+_16607122
|
0.007
|
ENST00000221671.3
ENST00000594035.1
ENST00000599550.1
ENST00000594813.1
|
C19orf44
|
chromosome 19 open reading frame 44
|
|
chr7_+_139044621
|
0.007
|
ENST00000354926.4
|
C7orf55-LUC7L2
|
C7orf55-LUC7L2 readthrough
|
|
chr12_-_120806960
|
0.007
|
ENST00000257552.2
|
MSI1
|
musashi RNA-binding protein 1
|
|
chr2_-_228028829
|
0.006
|
ENST00000396625.3
ENST00000329662.7
|
COL4A4
|
collagen, type IV, alpha 4
|
|
chr19_+_34919257
|
0.006
|
ENST00000246548.4
ENST00000590048.2
|
UBA2
|
ubiquitin-like modifier activating enzyme 2
|
|
chr10_+_102222798
|
0.006
|
ENST00000343737.5
|
WNT8B
|
wingless-type MMTV integration site family, member 8B
|
|
chr8_+_120220561
|
0.005
|
ENST00000276681.6
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr8_+_77593448
|
0.005
|
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr19_-_18392422
|
0.005
|
ENST00000252818.3
|
JUND
|
jun D proto-oncogene
|
|
chr1_+_15853308
|
0.005
|
ENST00000375838.1
ENST00000375847.3
ENST00000375849.1
|
DNAJC16
|
DnaJ (Hsp40) homolog, subfamily C, member 16
|
|
chr1_+_43148059
|
0.004
|
ENST00000321358.7
ENST00000332220.6
|
YBX1
|
Y box binding protein 1
|
|
chr3_+_16216137
|
0.004
|
ENST00000339732.5
|
GALNT15
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15
|
|
chr14_-_30396948
|
0.003
|
ENST00000331968.5
|
PRKD1
|
protein kinase D1
|
|
chr3_+_88199099
|
0.003
|
ENST00000486971.1
|
C3orf38
|
chromosome 3 open reading frame 38
|
|
chr9_-_100935043
|
0.003
|
ENST00000343933.5
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr15_+_41709302
|
0.002
|
ENST00000389629.4
|
RTF1
|
Rtf1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chrX_+_56259316
|
0.002
|
ENST00000468660.1
|
KLF8
|
Kruppel-like factor 8
|
|
chr17_-_617949
|
0.002
|
ENST00000401468.3
ENST00000574029.1
ENST00000291074.5
ENST00000571805.1
ENST00000437048.2
ENST00000446250.2
|
VPS53
|
vacuolar protein sorting 53 homolog (S. cerevisiae)
|
|
chr1_+_66999799
|
0.001
|
ENST00000371035.3
ENST00000371036.3
ENST00000371037.4
|
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1
|
|
chr3_+_152017181
|
0.000
|
ENST00000498502.1
ENST00000324196.5
ENST00000545754.1
ENST00000357472.3
|
MBNL1
|
muscleblind-like splicing regulator 1
|
|
chrX_+_154997474
|
0.000
|
ENST00000302805.2
|
SPRY3
|
sprouty homolog 3 (Drosophila)
|