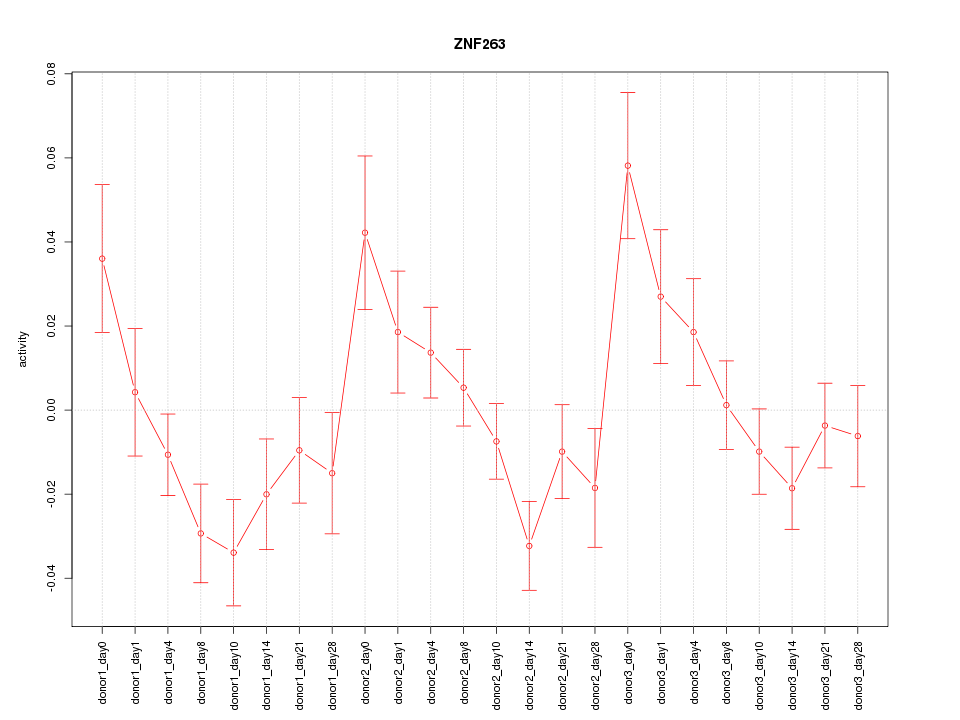
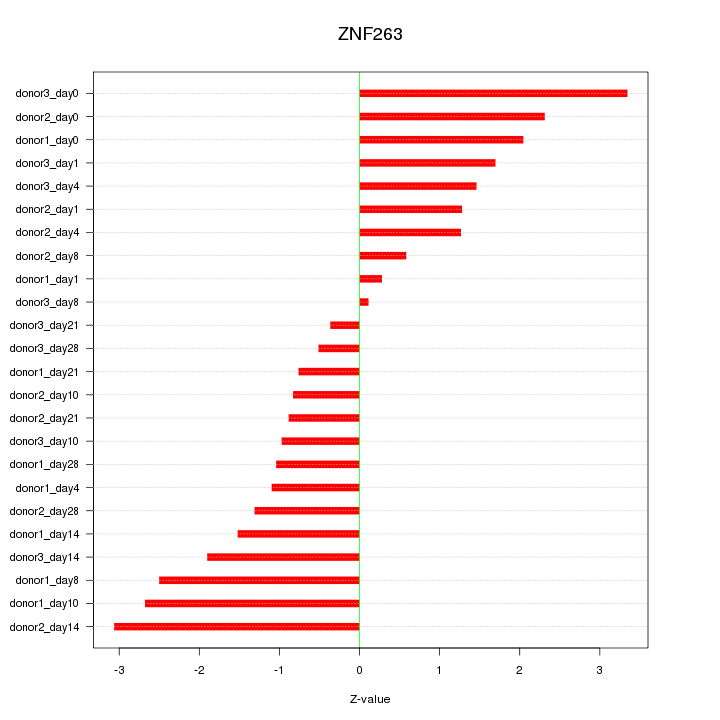
Motif ID: ZNF263
Z-value: 1.653
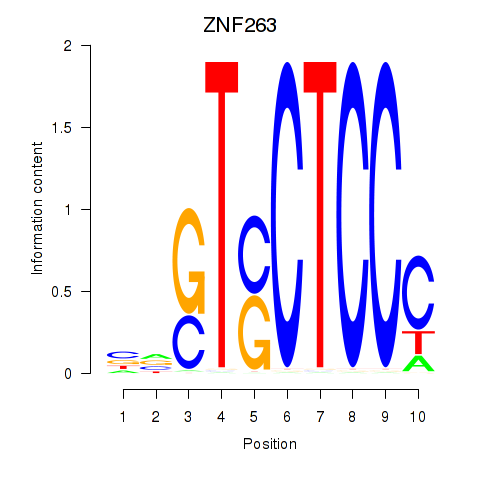
Transcription factors associated with ZNF263:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| ZNF263 | ENSG00000006194.6 | ZNF263 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF263 | hg19_v2_chr16_+_3313791_3313834 | -0.11 | 6.0e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 14.5 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 2.0 | 5.9 | GO:0060516 | primary prostatic bud elongation(GO:0060516) renal vesicle induction(GO:0072034) |
| 1.6 | 14.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 1.6 | 9.5 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 1.6 | 12.6 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 1.4 | 1.4 | GO:0060458 | right lung development(GO:0060458) |
| 1.3 | 3.8 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 1.2 | 3.5 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 1.1 | 6.4 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 1.0 | 3.0 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 1.0 | 2.9 | GO:0052553 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.9 | 3.7 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.9 | 2.8 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.9 | 2.8 | GO:0032499 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) detection of peptidoglycan(GO:0032499) |
| 0.9 | 9.9 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.9 | 5.4 | GO:0032252 | secretory granule localization(GO:0032252) endoplasmic reticulum localization(GO:0051643) |
| 0.9 | 4.5 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.9 | 2.7 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.9 | 2.6 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.8 | 17.8 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.8 | 4.7 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.8 | 8.4 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.7 | 2.2 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) interleukin-1 beta biosynthetic process(GO:0050720) response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.7 | 3.5 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.7 | 2.0 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.7 | 16.6 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.7 | 4.6 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.7 | 0.7 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.6 | 1.9 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.6 | 2.6 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.6 | 1.9 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.6 | 1.8 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.6 | 2.9 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.5 | 2.2 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.5 | 3.1 | GO:2000683 | regulation of cellular response to X-ray(GO:2000683) |
| 0.5 | 2.6 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.5 | 2.4 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.5 | 1.4 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.5 | 1.8 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.4 | 3.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.4 | 1.8 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.4 | 3.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.4 | 0.9 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.4 | 0.4 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.4 | 1.7 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.4 | 3.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.4 | 4.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.4 | 1.2 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.4 | 1.5 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.4 | 0.8 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.4 | 0.4 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.4 | 2.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.4 | 1.4 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.3 | 1.4 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.3 | 1.7 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.3 | 2.6 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.3 | 1.9 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.3 | 2.8 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 1.8 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.3 | 0.9 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.3 | 0.9 | GO:0050894 | determination of affect(GO:0050894) |
| 0.3 | 2.6 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 0.9 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.3 | 2.6 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.3 | 0.8 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.3 | 1.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.3 | 4.7 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.3 | 0.8 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.3 | 3.8 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.3 | 1.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.3 | 0.5 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.3 | 1.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.3 | 1.0 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.3 | 1.5 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.3 | 1.0 | GO:0019427 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.2 | 0.7 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) |
| 0.2 | 1.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.2 | 3.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.2 | 0.7 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.2 | 0.7 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.2 | 1.2 | GO:0048749 | compound eye development(GO:0048749) |
| 0.2 | 0.7 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.2 | 6.5 | GO:0034380 | high-density lipoprotein particle assembly(GO:0034380) |
| 0.2 | 1.2 | GO:0048066 | developmental pigmentation(GO:0048066) |
| 0.2 | 0.7 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.2 | 3.6 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 | 0.7 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.2 | 1.2 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.2 | 0.9 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.2 | 2.9 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.2 | 1.1 | GO:1903285 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 2.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.2 | 2.8 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 1.3 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.2 | 2.6 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.2 | 1.5 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.2 | 3.4 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.2 | 1.2 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.2 | 1.6 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.2 | 1.0 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.2 | 1.0 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.2 | 0.8 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.2 | 0.8 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.2 | 1.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 2.9 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 1.0 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.2 | 1.5 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 0.2 | GO:0051714 | positive regulation of cytolysis in other organism(GO:0051714) |
| 0.2 | 3.4 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.2 | 0.9 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.2 | 0.9 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.2 | 2.5 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 1.6 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.2 | 1.2 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.2 | 1.5 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 1.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 1.3 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.2 | 2.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 0.7 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.2 | 2.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 0.8 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.2 | 0.5 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.2 | 1.4 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.2 | 2.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.2 | 0.5 | GO:0002605 | negative regulation of dendritic cell antigen processing and presentation(GO:0002605) regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 0.2 | 4.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 2.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.7 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.4 | GO:0021586 | pons maturation(GO:0021586) superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.8 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 0.7 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.1 | 0.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.3 | GO:0051783 | regulation of nuclear division(GO:0051783) |
| 0.1 | 0.4 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.1 | 1.4 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 2.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.4 | GO:2000687 | signal transduction by trans-phosphorylation(GO:0023016) negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 1.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.6 | GO:0030047 | actin modification(GO:0030047) |
| 0.1 | 0.9 | GO:0045598 | regulation of fat cell differentiation(GO:0045598) |
| 0.1 | 0.7 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 0.2 | GO:0061053 | somite development(GO:0061053) |
| 0.1 | 0.6 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.7 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.1 | 6.5 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 0.5 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 0.6 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.6 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.8 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.1 | 0.1 | GO:1904170 | regulation of bleb assembly(GO:1904170) positive regulation of bleb assembly(GO:1904172) |
| 0.1 | 0.4 | GO:1900020 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 | 2.9 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.3 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.1 | 1.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.8 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.8 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.4 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.1 | 1.0 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.1 | 0.6 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 0.5 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.1 | 0.4 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.1 | 0.5 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.9 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 4.0 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 2.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.9 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.1 | 0.3 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.1 | 0.8 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.1 | 0.4 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.8 | GO:0015820 | leucine transport(GO:0015820) proline transmembrane transport(GO:0035524) |
| 0.1 | 0.4 | GO:0000189 | MAPK import into nucleus(GO:0000189) regulation of Golgi inheritance(GO:0090170) |
| 0.1 | 2.4 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 1.6 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 2.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.2 | GO:0072364 | regulation of cellular ketone metabolic process by regulation of transcription from RNA polymerase II promoter(GO:0072364) |
| 0.1 | 1.7 | GO:0071474 | cellular hyperosmotic response(GO:0071474) |
| 0.1 | 0.3 | GO:0045658 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.5 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.1 | 0.6 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 1.5 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 0.6 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.6 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.5 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.1 | 0.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.3 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 6.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.7 | GO:0030578 | PML body organization(GO:0030578) |
| 0.1 | 0.3 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 | 0.3 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 2.2 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 3.5 | GO:0007616 | long-term memory(GO:0007616) |
| 0.1 | 0.8 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.1 | 0.6 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.1 | 0.5 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.1 | 4.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 5.4 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 1.1 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 0.9 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 1.4 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.4 | GO:0009439 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.1 | 0.4 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 0.4 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.6 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.1 | 1.5 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 0.7 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.9 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.1 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.1 | 0.3 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 1.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.4 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 1.0 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.5 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.1 | 1.6 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 2.2 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.3 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 0.7 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.4 | GO:0043316 | cytotoxic T cell degranulation(GO:0043316) positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 1.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.2 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 | 0.3 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.4 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.4 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.2 | GO:1904640 | response to methionine(GO:1904640) |
| 0.1 | 0.5 | GO:1901725 | regulation of histone deacetylase activity(GO:1901725) |
| 0.1 | 0.6 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.3 | GO:0046856 | phospholipid dephosphorylation(GO:0046839) phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.3 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.6 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.1 | 3.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.3 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.1 | 0.9 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 1.3 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.1 | 0.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.1 | 4.0 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 0.5 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.1 | 0.5 | GO:0046078 | pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) dUMP metabolic process(GO:0046078) |
| 0.1 | 0.4 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 1.9 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.4 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.6 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.5 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.5 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.6 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 1.0 | GO:0030903 | notochord development(GO:0030903) |
| 0.1 | 0.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.6 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.1 | 1.0 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 0.4 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.6 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.8 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.1 | 0.5 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 0.5 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 1.1 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 1.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.6 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 0.9 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.9 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 0.2 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 0.1 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.1 | 0.8 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 3.9 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.1 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 1.0 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.8 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.3 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 2.3 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 1.3 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.1 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.5 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.7 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.8 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.7 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.4 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.3 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0050923 | chemorepulsion of branchiomotor axon(GO:0021793) regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 2.0 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.0 | 0.5 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 3.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.3 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.6 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.4 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.5 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.5 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 1.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 1.3 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.7 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.3 | GO:0042325 | regulation of phosphorylation(GO:0042325) |
| 0.0 | 0.3 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.2 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.2 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.3 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 1.2 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.5 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 1.8 | GO:0030516 | regulation of axon extension(GO:0030516) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.5 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.5 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.4 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.3 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.6 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.3 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.5 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.2 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 0.0 | 0.6 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.4 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.3 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.4 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.2 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.3 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.4 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 1.1 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.4 | GO:0042339 | keratan sulfate biosynthetic process(GO:0018146) keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.3 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 | 0.6 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.3 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.4 | GO:0051271 | negative regulation of cellular component movement(GO:0051271) |
| 0.0 | 0.4 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.2 | GO:0060148 | positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.2 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 1.0 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 1.6 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.4 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.0 | 0.1 | GO:1901536 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 1.0 | GO:0048512 | circadian behavior(GO:0048512) |
| 0.0 | 0.8 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.2 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.7 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 0.3 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.7 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.1 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.0 | 0.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 1.6 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.1 | GO:0009183 | ADP biosynthetic process(GO:0006172) purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.0 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.0 | GO:0010749 | regulation of nitric oxide mediated signal transduction(GO:0010749) |
| 0.0 | 0.4 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.3 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.3 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 1.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.6 | GO:0006582 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.2 | GO:0032962 | regulation of inositol trisphosphate biosynthetic process(GO:0032960) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.2 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.0 | GO:0000423 | macromitophagy(GO:0000423) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0060268 | negative regulation of respiratory burst(GO:0060268) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.4 | GO:0010629 | negative regulation of gene expression(GO:0010629) |
| 0.0 | 0.1 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.3 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.2 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.0 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.0 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.7 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 1.5 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0075713 | establishment of integrated proviral latency(GO:0075713) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.5 | GO:0008283 | cell proliferation(GO:0008283) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.2 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.4 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.1 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 | 0.7 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 0.1 | GO:1901908 | diadenosine polyphosphate metabolic process(GO:0015959) diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.0 | 0.2 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 0.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.4 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 1.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.3 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.2 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 1.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0072678 | T cell migration(GO:0072678) |
| 0.0 | 0.2 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.8 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.1 | GO:0097646 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 1.5 | GO:0034332 | adherens junction organization(GO:0034332) |
| 0.0 | 0.2 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.4 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.3 | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway(GO:0007169) |
| 0.0 | 0.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 1.4 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 1.5 | GO:0090002 | establishment of protein localization to plasma membrane(GO:0090002) |
| 0.0 | 0.6 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 14.5 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 2.1 | 6.2 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.9 | 2.8 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.9 | 3.7 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.9 | 5.4 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.9 | 2.6 | GO:0097453 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.8 | 13.7 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.6 | 5.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.6 | 15.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.6 | 3.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.6 | 5.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.5 | 2.9 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.4 | 3.5 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.4 | 2.3 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.4 | 1.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.3 | 3.5 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.3 | 1.4 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.3 | 2.7 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.3 | 2.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.3 | 1.5 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.3 | 2.0 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.3 | 4.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.3 | 4.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.3 | 3.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.2 | 0.9 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 1.2 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.2 | 0.9 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.2 | 2.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 5.9 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.2 | 1.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 0.6 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 2.2 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 0.5 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 0.7 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.6 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.1 | 0.4 | GO:0031970 | organelle envelope lumen(GO:0031970) |
| 0.1 | 0.9 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 1.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.8 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.1 | 3.7 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 2.2 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 1.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 1.0 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 14.0 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 1.0 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.4 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 4.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.4 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.3 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 0.7 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 1.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 1.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.3 | GO:0097059 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 1.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 7.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.2 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.1 | 1.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 2.0 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 0.2 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.1 | 1.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 0.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 1.8 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.1 | 0.4 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 0.6 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 0.5 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 1.0 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 2.3 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.6 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 1.0 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.1 | 0.2 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.4 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 3.6 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.4 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 8.0 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 4.0 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 1.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 6.6 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 1.1 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 2.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 3.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 4.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 3.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.4 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.5 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 2.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.6 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 2.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.3 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.9 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 1.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 5.4 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.3 | GO:0097346 | INO80-type complex(GO:0097346) |
| 0.0 | 2.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.4 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 10.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 5.9 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 1.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.3 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.7 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.2 | GO:0001725 | stress fiber(GO:0001725) actin filament bundle(GO:0032432) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 2.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.3 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 11.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 4.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.0 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 18.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 1.6 | 9.5 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 1.3 | 3.8 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 1.2 | 9.9 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 1.2 | 16.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 1.1 | 3.4 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.1 | 6.4 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 1.0 | 5.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.9 | 4.5 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.8 | 2.4 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.7 | 2.9 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.7 | 2.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.7 | 2.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.6 | 4.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.6 | 1.8 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.6 | 6.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.5 | 2.1 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.5 | 2.0 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.5 | 1.4 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.4 | 2.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.4 | 1.2 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.4 | 1.6 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.4 | 2.8 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.4 | 3.9 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.4 | 6.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.4 | 1.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.4 | 2.9 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.4 | 1.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.4 | 1.4 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.3 | 2.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.3 | 2.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.3 | 3.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.3 | 1.2 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.3 | 1.2 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.3 | 2.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.3 | 0.9 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.3 | 3.8 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.3 | 3.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.3 | 0.8 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.3 | 0.8 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.3 | 2.7 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.3 | 1.8 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.3 | 2.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 1.5 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.2 | 0.7 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.2 | 0.7 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 1.6 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 1.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 2.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 1.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.2 | 1.7 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 3.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 3.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 4.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 2.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 1.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.2 | 1.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.2 | 0.8 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.2 | 0.6 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.2 | 2.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.2 | 0.6 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 4.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 0.9 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.2 | 5.8 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.2 | 1.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 0.6 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.2 | 0.2 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.2 | 0.9 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.2 | 0.9 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 5.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 0.5 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.2 | 3.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 2.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 0.5 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.2 | 51.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 0.5 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.2 | 1.9 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.2 | 0.5 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.4 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.1 | 1.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.4 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 1.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 0.4 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 0.8 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.5 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 1.8 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.1 | 3.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.4 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.1 | 0.4 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.1 | 0.9 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 2.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 1.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.5 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.1 | 1.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 1.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.5 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 1.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.6 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 1.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.3 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.7 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.9 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 0.4 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.3 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 0.4 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.1 | 0.5 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 1.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.8 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.1 | 0.8 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.3 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.1 | 2.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.5 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 2.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.6 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 1.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 3.6 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.1 | 0.9 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.3 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.1 | 1.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.6 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 4.4 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 0.4 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 3.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.8 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 2.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 1.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.7 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.0 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 3.4 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 0.3 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.2 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 0.6 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 0.4 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.5 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.6 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 1.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.5 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.1 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.3 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.3 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 0.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.6 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 1.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.4 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 1.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 7.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 2.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.5 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.8 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 0.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 6.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.5 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 2.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 5.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 5.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.3 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.5 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.9 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 2.2 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 3.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 4.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.2 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.3 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 1.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 1.0 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 1.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 1.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.8 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.5 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.8 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 3.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.2 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 0.5 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.3 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.6 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.3 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.8 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.4 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 5.2 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 3.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.5 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 6.9 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.5 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.5 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 4.6 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 0.7 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0044388 | ubiquitin activating enzyme activity(GO:0004839) small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.3 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0019209 | kinase activator activity(GO:0019209) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 2.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.7 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 7.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.0 | 0.4 | GO:0042166 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.0 | 0.5 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 1.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 2.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 1.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.0 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.7 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.9 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.7 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 18.2 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.3 | 14.5 | PID_SYNDECAN_2_PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 6.7 | PID_ANTHRAX_PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 6.1 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.2 | 2.5 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.2 | 10.3 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 6.6 | PID_IL8_CXCR2_PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 5.6 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 0.9 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.4 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.1 | 4.4 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.1 | 6.1 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.1 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.5 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 5.1 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.1 | 2.1 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.1 | 4.3 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 1.9 | PID_EPHB_FWD_PATHWAY | EPHB forward signaling |
| 0.1 | 3.0 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.3 | PID_IL2_1PATHWAY | IL2-mediated signaling events |
| 0.1 | 1.6 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.3 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.1 | 1.1 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.1 | 17.3 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 5.8 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 2.0 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 3.3 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 0.9 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 0.3 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 0.8 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.1 | 1.5 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 1.8 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID_VEGFR1_PATHWAY | VEGFR1 specific signals |
| 0.0 | 5.1 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.3 | PID_NECTIN_PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.1 | PID_WNT_CANONICAL_PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.7 | PID_TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 13.2 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | PID_IL27_PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.4 | PID_ENDOTHELIN_PATHWAY | Endothelins |
| 0.0 | 2.2 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.9 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.1 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.0 | PID_IL1_PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.3 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.5 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.3 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.0 | 0.5 | PID_BCR_5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.6 | SIG_CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.6 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID_NEPHRIN_NEPH1_PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.1 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.1 | PID_AR_NONGENOMIC_PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.4 | ST_GA13_PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.5 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.0 | 2.8 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.1 | PID_TRAIL_PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.7 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.3 | SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.3 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.6 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.8 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.0 | 0.2 | PID_IFNG_PATHWAY | IFN-gamma pathway |
| 0.0 | 0.5 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.7 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID_PTP1B_PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.6 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.3 | REACTOME_REVERSIBLE_HYDRATION_OF_CARBON_DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.5 | 9.5 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 10.3 | REACTOME_HDL_MEDIATED_LIPID_TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 5.7 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.3 | 20.0 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 6.4 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 3.7 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 4.5 | REACTOME_SIGNAL_ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 9.9 | REACTOME_SEMA4D_IN_SEMAPHORIN_SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.2 | 3.0 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 3.6 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 1.7 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 4.6 | REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 12.5 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.2 | 3.1 | REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 0.8 | REACTOME_NEGATIVE_REGULATION_OF_THE_PI3K_AKT_NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 1.8 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 7.5 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.1 | 2.1 | REACTOME_RECYCLING_PATHWAY_OF_L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 4.3 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 3.3 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.1 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 3.1 | REACTOME_GRB2_SOS_PROVIDES_LINKAGE_TO_MAPK_SIGNALING_FOR_INTERGRINS_ | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 4.2 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 4.0 | REACTOME_STEROID_HORMONES | Genes involved in Steroid hormones |
| 0.1 | 3.1 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.7 | REACTOME_INFLUENZA_LIFE_CYCLE | Genes involved in Influenza Life Cycle |
| 0.1 | 3.5 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_ACTIVATES_SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.1 | 1.0 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 3.0 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.7 | REACTOME_SIGNALLING_BY_NGF | Genes involved in Signalling by NGF |
| 0.1 | 3.1 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.1 | 1.6 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.1 | 1.6 | REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.1 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELL_ADHESION_PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.0 | REACTOME_ETHANOL_OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.6 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 4.4 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 0.8 | REACTOME_GROWTH_HORMONE_RECEPTOR_SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 3.3 | REACTOME_SIGNAL_TRANSDUCTION_BY_L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 1.1 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.0 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.1 | REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.3 | REACTOME_RAS_ACTIVATION_UOPN_CA2_INFUX_THROUGH_NMDA_RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 3.1 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.3 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.5 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.1 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 4.4 | REACTOME_CELL_JUNCTION_ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.5 | REACTOME_ABACAVIR_TRANSPORT_AND_METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.7 | REACTOME_INHIBITION_OF_INSULIN_SECRETION_BY_ADRENALINE_NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.5 | REACTOME_PLATELET_CALCIUM_HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.5 | REACTOME_EXTRACELLULAR_MATRIX_ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 1.3 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.3 | REACTOME_MUSCLE_CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.1 | REACTOME_DOPAMINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME_JNK_C_JUN_KINASES_PHOSPHORYLATION_AND_ACTIVATION_MEDIATED_BY_ACTIVATED_HUMAN_TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.8 | REACTOME_THE_ROLE_OF_NEF_IN_HIV1_REPLICATION_AND_DISEASE_PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.9 | REACTOME_FORMATION_OF_INCISION_COMPLEX_IN_GG_NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.6 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.4 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.0 | 2.7 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.9 | REACTOME_KERATAN_SULFATE_BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.9 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.9 | REACTOME_PHASE1_FUNCTIONALIZATION_OF_COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.4 | REACTOME_RNA_POL_III_CHAIN_ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.4 | REACTOME_SIGNALING_BY_ACTIVATED_POINT_MUTANTS_OF_FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.1 | REACTOME_DSCAM_INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 2.5 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.1 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.8 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.9 | REACTOME_INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.7 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.7 | REACTOME_CYTOSOLIC_SULFONATION_OF_SMALL_MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.7 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.0 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME_SIGNAL_REGULATORY_PROTEIN_SIRP_FAMILY_INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME_PROLACTIN_RECEPTOR_SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.9 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.6 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.6 | REACTOME_IL1_SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.5 | REACTOME_METABOLISM_OF_NON_CODING_RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.7 | REACTOME_GAB1_SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.0 | 1.0 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.8 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.1 | REACTOME_CTNNB1_PHOSPHORYLATION_CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 2.2 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.5 | REACTOME_PPARA_ACTIVATES_GENE_EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.2 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.6 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME_DESTABILIZATION_OF_MRNA_BY_KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.3 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.9 | REACTOME_G_ALPHA1213_SIGNALLING_EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.1 | REACTOME_REGULATION_OF_ORNITHINE_DECARBOXYLASE_ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.4 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.2 | REACTOME_OLFACTORY_SIGNALING_PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.4 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME_INTEGRATION_OF_PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |