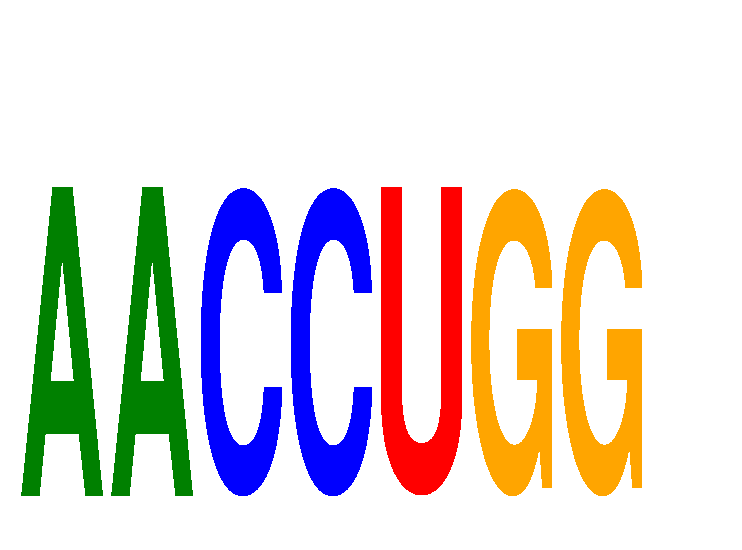Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for AACCUGG
Z-value: 0.09
miRNA associated with seed AACCUGG
| Name | miRBASE accession |
|---|---|
|
mmu-miR-490-3p
|
MIMAT0003780 |
Activity profile of AACCUGG motif
Sorted Z-values of AACCUGG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_42149288 | 0.10 |
ENSMUST00000115073.2
ENSMUST00000115072.1 |
Stag2
|
stromal antigen 2 |
| chr2_+_79255500 | 0.09 |
ENSMUST00000099972.4
|
Itga4
|
integrin alpha 4 |
| chr15_+_103240405 | 0.08 |
ENSMUST00000036004.9
ENSMUST00000087351.7 |
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr2_+_75659253 | 0.07 |
ENSMUST00000111964.1
ENSMUST00000111962.1 ENSMUST00000111961.1 ENSMUST00000164947.2 ENSMUST00000090792.4 |
Hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr1_-_143702832 | 0.06 |
ENSMUST00000018337.7
|
Cdc73
|
cell division cycle 73, Paf1/RNA polymerase II complex component |
| chr6_-_39206782 | 0.06 |
ENSMUST00000002305.8
|
Jhdm1d
|
jumonji C domain-containing histone demethylase 1 homolog D (S. cerevisiae) |
| chr2_-_37647199 | 0.03 |
ENSMUST00000028279.3
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr5_-_20951769 | 0.03 |
ENSMUST00000036489.5
|
Rsbn1l
|
round spermatid basic protein 1-like |
| chr13_+_41606216 | 0.03 |
ENSMUST00000129449.1
|
Tmem170b
|
transmembrane protein 170B |
| chr5_+_108268897 | 0.02 |
ENSMUST00000031190.4
|
Dr1
|
down-regulator of transcription 1 |
| chr6_-_51469869 | 0.02 |
ENSMUST00000114459.1
ENSMUST00000069949.6 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr15_+_54410755 | 0.02 |
ENSMUST00000036737.3
|
Colec10
|
collectin sub-family member 10 |
| chr7_-_98309471 | 0.02 |
ENSMUST00000033020.7
|
Acer3
|
alkaline ceramidase 3 |
| chr14_+_118854695 | 0.02 |
ENSMUST00000100314.3
|
Cldn10
|
claudin 10 |
| chr5_-_21055880 | 0.02 |
ENSMUST00000030556.7
|
Ptpn12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr6_-_71440623 | 0.02 |
ENSMUST00000002292.8
|
Rmnd5a
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| chr5_+_88720855 | 0.01 |
ENSMUST00000113229.1
ENSMUST00000006424.7 |
Mob1b
|
MOB kinase activator 1B |
| chr6_-_52234902 | 0.01 |
ENSMUST00000125581.1
|
Hoxa10
|
homeobox A10 |
| chr4_+_34550937 | 0.01 |
ENSMUST00000084299.5
|
Akirin2
|
akirin 2 |
| chr8_-_47990535 | 0.01 |
ENSMUST00000057561.7
|
Wwc2
|
WW, C2 and coiled-coil domain containing 2 |
| chr9_+_72532214 | 0.01 |
ENSMUST00000163401.2
ENSMUST00000093820.3 |
Rfx7
|
regulatory factor X, 7 |
| chr10_+_75518042 | 0.01 |
ENSMUST00000020397.8
|
Snrpd3
|
small nuclear ribonucleoprotein D3 |
| chr5_+_72914264 | 0.01 |
ENSMUST00000144843.1
|
Slain2
|
SLAIN motif family, member 2 |
| chr2_-_174472949 | 0.01 |
ENSMUST00000016401.8
|
Slmo2
|
slowmo homolog 2 (Drosophila) |
| chr6_-_38876163 | 0.01 |
ENSMUST00000161779.1
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr17_+_64600702 | 0.01 |
ENSMUST00000086723.3
|
Man2a1
|
mannosidase 2, alpha 1 |
| chr18_-_79109391 | 0.01 |
ENSMUST00000025430.8
ENSMUST00000161465.2 |
Setbp1
|
SET binding protein 1 |
| chr17_+_24414640 | 0.00 |
ENSMUST00000115371.1
ENSMUST00000088512.5 ENSMUST00000163717.1 |
Rnps1
|
ribonucleic acid binding protein S1 |
| chr18_+_60526194 | 0.00 |
ENSMUST00000025505.5
|
Dctn4
|
dynactin 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AACCUGG
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |