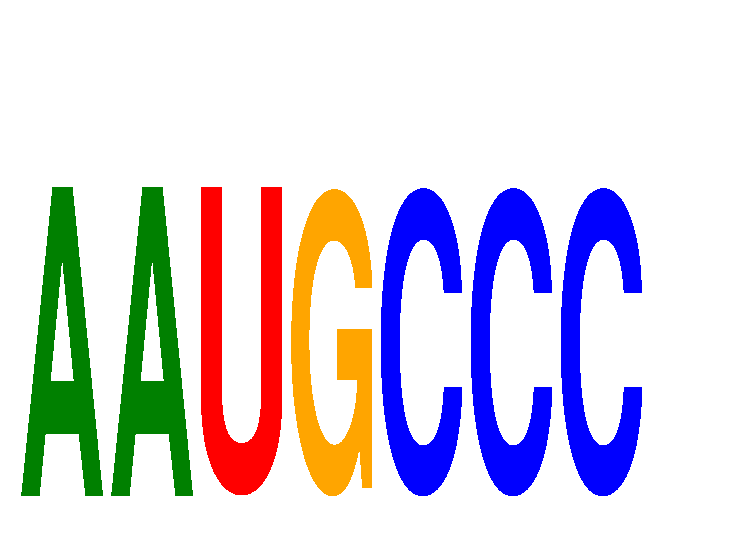Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for AAUGCCC
Z-value: 0.53
miRNA associated with seed AAUGCCC
| Name | miRBASE accession |
|---|---|
|
mmu-miR-365-3p
|
MIMAT0000711 |
Activity profile of AAUGCCC motif
Sorted Z-values of AAUGCCC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_87859255 | 1.69 |
ENSMUST00000105300.2
|
Igf1
|
insulin-like growth factor 1 |
| chr10_-_114801364 | 1.66 |
ENSMUST00000061632.7
|
Trhde
|
TRH-degrading enzyme |
| chr8_+_76899772 | 1.61 |
ENSMUST00000109913.2
|
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr14_-_18239053 | 1.55 |
ENSMUST00000090543.5
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr6_-_138043411 | 1.27 |
ENSMUST00000111873.1
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr1_-_105356658 | 0.91 |
ENSMUST00000058688.5
ENSMUST00000172299.1 |
Rnf152
|
ring finger protein 152 |
| chr9_+_108692116 | 0.90 |
ENSMUST00000035220.6
|
Prkar2a
|
protein kinase, cAMP dependent regulatory, type II alpha |
| chr11_+_28853189 | 0.90 |
ENSMUST00000020759.5
|
Efemp1
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 1 |
| chr4_-_19708922 | 0.88 |
ENSMUST00000108246.2
|
Wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr15_+_41789495 | 0.81 |
ENSMUST00000090095.5
ENSMUST00000022918.7 |
Oxr1
|
oxidation resistance 1 |
| chr5_-_106696819 | 0.79 |
ENSMUST00000127434.1
ENSMUST00000112696.1 ENSMUST00000112698.1 |
Zfp644
|
zinc finger protein 644 |
| chr2_-_53191214 | 0.79 |
ENSMUST00000076313.6
ENSMUST00000125243.1 |
Prpf40a
|
PRP40 pre-mRNA processing factor 40 homolog A (yeast) |
| chr4_+_100095791 | 0.79 |
ENSMUST00000039630.5
|
Ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr3_-_121815212 | 0.75 |
ENSMUST00000029770.5
|
Abcd3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr2_+_71981184 | 0.69 |
ENSMUST00000090826.5
ENSMUST00000102698.3 |
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr15_+_58415456 | 0.63 |
ENSMUST00000037270.3
|
D15Ertd621e
|
DNA segment, Chr 15, ERATO Doi 621, expressed |
| chr17_+_3326552 | 0.61 |
ENSMUST00000169838.1
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr5_+_63649335 | 0.59 |
ENSMUST00000159584.1
|
3110047P20Rik
|
RIKEN cDNA 3110047P20 gene |
| chr13_-_64274879 | 0.57 |
ENSMUST00000109770.1
|
Cdc14b
|
CDC14 cell division cycle 14B |
| chr2_+_55437100 | 0.57 |
ENSMUST00000112633.2
ENSMUST00000112632.1 |
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr5_-_66173051 | 0.54 |
ENSMUST00000113726.1
|
Rbm47
|
RNA binding motif protein 47 |
| chr19_-_29805989 | 0.51 |
ENSMUST00000177155.1
ENSMUST00000059484.7 |
9930021J03Rik
|
RIKEN cDNA 9930021J03 gene |
| chr2_-_103303179 | 0.50 |
ENSMUST00000090475.3
|
Ehf
|
ets homologous factor |
| chr4_+_41465134 | 0.49 |
ENSMUST00000030154.6
|
Nudt2
|
nudix (nucleoside diphosphate linked moiety X)-type motif 2 |
| chr10_+_39732099 | 0.48 |
ENSMUST00000019986.6
|
Rev3l
|
REV3-like, catalytic subunit of DNA polymerase zeta RAD54 like (S. cerevisiae) |
| chr10_+_99108135 | 0.47 |
ENSMUST00000161240.2
|
Galnt4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 |
| chr14_-_39472825 | 0.45 |
ENSMUST00000168810.2
ENSMUST00000173780.1 ENSMUST00000166968.2 |
Nrg3
|
neuregulin 3 |
| chr18_-_39490649 | 0.44 |
ENSMUST00000115567.1
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr16_+_34784917 | 0.44 |
ENSMUST00000023538.8
|
Mylk
|
myosin, light polypeptide kinase |
| chr3_+_152346465 | 0.43 |
ENSMUST00000026507.6
ENSMUST00000117492.2 |
Usp33
|
ubiquitin specific peptidase 33 |
| chr12_+_40446050 | 0.43 |
ENSMUST00000037488.6
|
Dock4
|
dedicator of cytokinesis 4 |
| chr13_-_8871696 | 0.42 |
ENSMUST00000054251.6
ENSMUST00000176813.1 |
Wdr37
|
WD repeat domain 37 |
| chr1_-_13589717 | 0.41 |
ENSMUST00000027068.4
|
Tram1
|
translocating chain-associating membrane protein 1 |
| chr17_+_26715644 | 0.39 |
ENSMUST00000062519.7
ENSMUST00000144221.1 ENSMUST00000142539.1 ENSMUST00000151681.1 |
Crebrf
|
CREB3 regulatory factor |
| chr10_+_84576626 | 0.39 |
ENSMUST00000020223.7
|
Tcp11l2
|
t-complex 11 (mouse) like 2 |
| chr19_-_6921804 | 0.38 |
ENSMUST00000025906.4
|
Esrra
|
estrogen related receptor, alpha |
| chr8_+_90828820 | 0.38 |
ENSMUST00000109614.2
ENSMUST00000048665.6 |
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr10_+_86300372 | 0.37 |
ENSMUST00000020234.7
|
Timp3
|
tissue inhibitor of metalloproteinase 3 |
| chr6_+_21949571 | 0.37 |
ENSMUST00000031680.3
ENSMUST00000115389.1 ENSMUST00000151473.1 |
Ing3
|
inhibitor of growth family, member 3 |
| chr9_-_75559604 | 0.36 |
ENSMUST00000072232.7
|
Tmod3
|
tropomodulin 3 |
| chrX_-_104413825 | 0.36 |
ENSMUST00000033695.5
|
Abcb7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr14_-_31830402 | 0.36 |
ENSMUST00000014640.7
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr19_+_32757497 | 0.34 |
ENSMUST00000013807.7
|
Pten
|
phosphatase and tensin homolog |
| chr5_+_23434435 | 0.34 |
ENSMUST00000094962.2
ENSMUST00000115128.1 |
Kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chr1_-_64737735 | 0.34 |
ENSMUST00000063982.5
ENSMUST00000116133.2 |
Fzd5
|
frizzled homolog 5 (Drosophila) |
| chr2_-_140066661 | 0.33 |
ENSMUST00000046656.2
ENSMUST00000099304.3 ENSMUST00000110079.2 |
Tasp1
|
taspase, threonine aspartase 1 |
| chr10_+_106470281 | 0.33 |
ENSMUST00000029404.9
ENSMUST00000169303.1 |
Ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr9_-_101251810 | 0.32 |
ENSMUST00000075941.5
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr16_-_11203259 | 0.31 |
ENSMUST00000119953.1
|
Rsl1d1
|
ribosomal L1 domain containing 1 |
| chr16_-_30388530 | 0.30 |
ENSMUST00000100013.2
ENSMUST00000061350.6 |
Atp13a3
|
ATPase type 13A3 |
| chr4_+_116221633 | 0.29 |
ENSMUST00000030464.7
|
Pik3r3
|
phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 3 (p55) |
| chr6_-_114921778 | 0.29 |
ENSMUST00000032459.7
|
Vgll4
|
vestigial like 4 (Drosophila) |
| chr17_+_86753900 | 0.28 |
ENSMUST00000024954.9
|
Epas1
|
endothelial PAS domain protein 1 |
| chr5_+_110135823 | 0.28 |
ENSMUST00000112519.2
ENSMUST00000014812.8 |
Chfr
|
checkpoint with forkhead and ring finger domains |
| chr19_-_47464406 | 0.27 |
ENSMUST00000111800.2
ENSMUST00000081619.2 |
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr17_-_80480435 | 0.27 |
ENSMUST00000068714.5
|
Sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr4_-_132212255 | 0.27 |
ENSMUST00000152796.1
|
Ythdf2
|
YTH domain family 2 |
| chr2_+_109890846 | 0.26 |
ENSMUST00000028583.7
|
Lin7c
|
lin-7 homolog C (C. elegans) |
| chr5_-_24842579 | 0.26 |
ENSMUST00000030787.8
|
Rheb
|
Ras homolog enriched in brain |
| chr2_-_12301914 | 0.26 |
ENSMUST00000028106.4
|
Itga8
|
integrin alpha 8 |
| chr11_+_33963013 | 0.25 |
ENSMUST00000020362.2
|
Kcnmb1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
| chr11_-_62457289 | 0.24 |
ENSMUST00000069456.4
ENSMUST00000018645.6 |
Ncor1
|
nuclear receptor co-repressor 1 |
| chr11_-_51857624 | 0.22 |
ENSMUST00000020655.7
ENSMUST00000109090.1 |
Phf15
|
PHD finger protein 15 |
| chr6_-_88446491 | 0.21 |
ENSMUST00000165242.1
|
Eefsec
|
eukaryotic elongation factor, selenocysteine-tRNA-specific |
| chr11_-_104442232 | 0.20 |
ENSMUST00000106977.1
ENSMUST00000106972.1 |
Kansl1
|
KAT8 regulatory NSL complex subunit 1 |
| chr10_-_56228636 | 0.20 |
ENSMUST00000099739.3
|
Tbc1d32
|
TBC1 domain family, member 32 |
| chr6_+_115931922 | 0.19 |
ENSMUST00000032471.6
|
Rho
|
rhodopsin |
| chr2_-_48949206 | 0.19 |
ENSMUST00000090976.3
ENSMUST00000149679.1 ENSMUST00000028098.4 |
Orc4
|
origin recognition complex, subunit 4 |
| chr14_-_47568427 | 0.19 |
ENSMUST00000042988.6
|
Atg14
|
autophagy related 14 |
| chr6_-_99666762 | 0.19 |
ENSMUST00000032151.2
|
Eif4e3
|
eukaryotic translation initiation factor 4E member 3 |
| chr6_-_116193426 | 0.19 |
ENSMUST00000088896.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr15_+_102406143 | 0.19 |
ENSMUST00000170884.1
ENSMUST00000165924.1 ENSMUST00000163709.1 ENSMUST00000001326.6 |
Sp1
|
trans-acting transcription factor 1 |
| chr10_-_118868903 | 0.18 |
ENSMUST00000004281.8
|
Dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr7_-_17027853 | 0.18 |
ENSMUST00000003183.5
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr1_-_171150588 | 0.17 |
ENSMUST00000155798.1
ENSMUST00000081560.4 ENSMUST00000111336.3 |
Sdhc
|
succinate dehydrogenase complex, subunit C, integral membrane protein |
| chrX_-_103821940 | 0.17 |
ENSMUST00000042664.5
|
Slc16a2
|
solute carrier family 16 (monocarboxylic acid transporters), member 2 |
| chr9_+_108953578 | 0.17 |
ENSMUST00000112070.1
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr1_+_178798438 | 0.17 |
ENSMUST00000160789.1
|
Kif26b
|
kinesin family member 26B |
| chr8_+_111536492 | 0.17 |
ENSMUST00000168428.1
ENSMUST00000171182.1 |
Znrf1
|
zinc and ring finger 1 |
| chr13_+_40704005 | 0.16 |
ENSMUST00000069457.1
|
Gm9979
|
predicted gene 9979 |
| chr9_-_57645561 | 0.16 |
ENSMUST00000034863.6
|
Csk
|
c-src tyrosine kinase |
| chr11_-_53423123 | 0.16 |
ENSMUST00000036045.5
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr19_-_60226666 | 0.15 |
ENSMUST00000065286.1
|
D19Ertd737e
|
DNA segment, Chr 19, ERATO Doi 737, expressed |
| chr2_+_18677002 | 0.15 |
ENSMUST00000028071.6
|
Bmi1
|
Bmi1 polycomb ring finger oncogene |
| chr12_-_56535047 | 0.14 |
ENSMUST00000178477.2
|
Nkx2-1
|
NK2 homeobox 1 |
| chr8_+_25602236 | 0.13 |
ENSMUST00000146919.1
ENSMUST00000142395.1 ENSMUST00000139966.1 |
Whsc1l1
|
Wolf-Hirschhorn syndrome candidate 1-like 1 (human) |
| chr3_+_135438722 | 0.13 |
ENSMUST00000166033.1
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr7_+_97579868 | 0.13 |
ENSMUST00000042399.7
ENSMUST00000107153.1 |
Rsf1
|
remodeling and spacing factor 1 |
| chr13_+_47122719 | 0.13 |
ENSMUST00000068891.4
|
Rnf144b
|
ring finger protein 144B |
| chr1_-_56972437 | 0.13 |
ENSMUST00000042857.7
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr16_+_33380765 | 0.13 |
ENSMUST00000165418.1
|
Zfp148
|
zinc finger protein 148 |
| chr2_+_105668888 | 0.13 |
ENSMUST00000111086.4
ENSMUST00000111087.3 |
Pax6
|
paired box gene 6 |
| chr13_+_80883403 | 0.12 |
ENSMUST00000099356.2
|
Arrdc3
|
arrestin domain containing 3 |
| chr15_+_58933774 | 0.11 |
ENSMUST00000022980.3
|
Ndufb9
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9 |
| chr10_-_121586730 | 0.11 |
ENSMUST00000020316.2
|
Tbk1
|
TANK-binding kinase 1 |
| chr16_-_60605226 | 0.10 |
ENSMUST00000068860.6
|
Epha6
|
Eph receptor A6 |
| chr15_-_93275151 | 0.10 |
ENSMUST00000057896.4
ENSMUST00000049484.6 |
Gxylt1
|
glucoside xylosyltransferase 1 |
| chr8_-_122974793 | 0.10 |
ENSMUST00000098334.5
|
Ankrd11
|
ankyrin repeat domain 11 |
| chr7_-_70360593 | 0.10 |
ENSMUST00000032768.7
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr1_+_34849950 | 0.09 |
ENSMUST00000027297.4
|
Plekhb2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr9_+_7184514 | 0.09 |
ENSMUST00000034499.8
|
Dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr5_-_96161990 | 0.09 |
ENSMUST00000155901.1
|
Cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chr1_+_131910458 | 0.09 |
ENSMUST00000062264.6
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr10_-_128645965 | 0.09 |
ENSMUST00000133342.1
|
Ikzf4
|
IKAROS family zinc finger 4 |
| chr13_-_96670838 | 0.09 |
ENSMUST00000022176.8
|
Hmgcr
|
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
| chr5_+_144100387 | 0.09 |
ENSMUST00000041804.7
|
Lmtk2
|
lemur tyrosine kinase 2 |
| chr16_+_21891969 | 0.09 |
ENSMUST00000042065.6
|
Map3k13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr15_-_98871175 | 0.09 |
ENSMUST00000178486.2
ENSMUST00000023741.9 |
Kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr9_+_72532214 | 0.08 |
ENSMUST00000163401.2
ENSMUST00000093820.3 |
Rfx7
|
regulatory factor X, 7 |
| chr11_+_88047693 | 0.07 |
ENSMUST00000079866.4
|
Srsf1
|
serine/arginine-rich splicing factor 1 |
| chr14_+_8080315 | 0.06 |
ENSMUST00000023924.3
|
Rpp14
|
ribonuclease P 14 subunit |
| chr4_+_123201503 | 0.06 |
ENSMUST00000068262.5
|
Nt5c1a
|
5'-nucleotidase, cytosolic IA |
| chr18_-_61400363 | 0.06 |
ENSMUST00000063307.5
ENSMUST00000075299.6 |
Ppargc1b
|
peroxisome proliferative activated receptor, gamma, coactivator 1 beta |
| chr11_-_119228461 | 0.05 |
ENSMUST00000036113.3
|
Tbc1d16
|
TBC1 domain family, member 16 |
| chr11_+_77216180 | 0.05 |
ENSMUST00000037912.5
ENSMUST00000156488.1 |
Ssh2
|
slingshot homolog 2 (Drosophila) |
| chr1_+_9798123 | 0.05 |
ENSMUST00000168907.1
ENSMUST00000166384.1 |
Sgk3
|
serum/glucocorticoid regulated kinase 3 |
| chr4_+_137594140 | 0.05 |
ENSMUST00000105840.1
ENSMUST00000055131.6 ENSMUST00000105839.1 ENSMUST00000105838.1 |
Usp48
|
ubiquitin specific peptidase 48 |
| chr19_+_6306456 | 0.04 |
ENSMUST00000025681.7
|
Cdc42bpg
|
CDC42 binding protein kinase gamma (DMPK-like) |
| chr6_-_22356068 | 0.04 |
ENSMUST00000163963.1
ENSMUST00000165576.1 |
Fam3c
|
family with sequence similarity 3, member C |
| chr10_-_112928974 | 0.04 |
ENSMUST00000099276.2
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr18_-_77713978 | 0.04 |
ENSMUST00000074653.4
|
8030462N17Rik
|
RIKEN cDNA 8030462N17 gene |
| chr7_-_47008397 | 0.04 |
ENSMUST00000061639.7
|
Spty2d1
|
SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae) |
| chr6_-_56797637 | 0.04 |
ENSMUST00000114323.1
|
Kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr15_+_6708372 | 0.03 |
ENSMUST00000061656.6
|
Rictor
|
RPTOR independent companion of MTOR, complex 2 |
| chr2_+_173659760 | 0.03 |
ENSMUST00000029024.3
|
Rab22a
|
RAB22A, member RAS oncogene family |
| chr11_+_20543307 | 0.03 |
ENSMUST00000093292.4
|
Sertad2
|
SERTA domain containing 2 |
| chr10_+_21994666 | 0.03 |
ENSMUST00000020145.5
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr17_+_87107621 | 0.02 |
ENSMUST00000041369.6
|
Socs5
|
suppressor of cytokine signaling 5 |
| chr4_+_63544747 | 0.02 |
ENSMUST00000035301.6
|
Atp6v1g1
|
ATPase, H+ transporting, lysosomal V1 subunit G1 |
| chr4_-_126429501 | 0.02 |
ENSMUST00000069097.6
|
Ago3
|
argonaute RISC catalytic subunit 3 |
| chr11_+_77686155 | 0.01 |
ENSMUST00000100802.4
ENSMUST00000181023.1 |
Nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr2_+_48814109 | 0.01 |
ENSMUST00000063886.3
|
Acvr2a
|
activin receptor IIA |
| chr4_-_82505749 | 0.01 |
ENSMUST00000107245.2
ENSMUST00000107246.1 |
Nfib
|
nuclear factor I/B |
| chr9_+_113930934 | 0.01 |
ENSMUST00000084885.5
ENSMUST00000009885.7 |
Ubp1
|
upstream binding protein 1 |
| chr1_-_91931172 | 0.00 |
ENSMUST00000097644.2
|
Hdac4
|
histone deacetylase 4 |
| chr7_+_5080214 | 0.00 |
ENSMUST00000098845.3
ENSMUST00000146317.1 ENSMUST00000153169.1 ENSMUST00000045277.6 |
Epn1
|
epsin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AAUGCCC
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.3 | 1.7 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.3 | 0.8 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 0.7 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.2 | 0.7 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.8 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 | 0.3 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.1 | 0.4 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.6 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.5 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.1 | 0.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 1.5 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.3 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.1 | 0.6 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.9 | GO:1903975 | regulation of glial cell migration(GO:1903975) |
| 0.1 | 0.2 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 | 0.3 | GO:1903179 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.1 | 0.9 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.3 | GO:1903677 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.1 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 0.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.3 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.0 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 1.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.1 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.2 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.3 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.2 | GO:0090005 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.1 | GO:0021918 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.0 | 0.8 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.7 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.3 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.8 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.1 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.7 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 0.5 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.2 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.8 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.2 | 0.9 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 1.6 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.8 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.4 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.9 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.7 | GO:0043539 | insulin-like growth factor receptor binding(GO:0005159) protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 1.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.5 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.2 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.9 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | SIG CHEMOTAXIS | Genes related to chemotaxis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.1 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.2 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |