Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
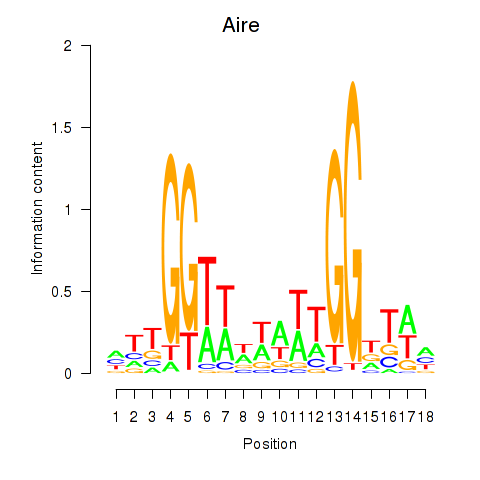
Results for Aire
Z-value: 0.65
Transcription factors associated with Aire
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Aire
|
ENSMUSG00000000731.9 | autoimmune regulator (autoimmune polyendocrinopathy candidiasis ectodermal dystrophy) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Aire | mm10_v2_chr10_-_78043580_78043611 | -0.04 | 8.3e-01 | Click! |
Activity profile of Aire motif
Sorted Z-values of Aire motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_39287074 | 2.57 |
ENSMUST00000003137.8
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr4_+_44943727 | 2.20 |
ENSMUST00000154177.1
|
Gm12678
|
predicted gene 12678 |
| chr5_+_130369420 | 2.07 |
ENSMUST00000086029.3
|
Caln1
|
calneuron 1 |
| chr5_-_87091150 | 1.74 |
ENSMUST00000154455.1
|
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr12_+_84052114 | 1.53 |
ENSMUST00000120927.1
|
Acot3
|
acyl-CoA thioesterase 3 |
| chr1_+_88200601 | 1.46 |
ENSMUST00000049289.8
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
| chr6_+_42245907 | 1.42 |
ENSMUST00000031897.5
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr8_-_25038875 | 1.28 |
ENSMUST00000084031.4
|
Htra4
|
HtrA serine peptidase 4 |
| chr19_+_3323301 | 1.18 |
ENSMUST00000025835.4
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr12_-_104865076 | 1.16 |
ENSMUST00000109937.1
ENSMUST00000109936.1 |
Clmn
|
calmin |
| chr10_-_107123585 | 1.10 |
ENSMUST00000165067.1
ENSMUST00000044668.4 |
Acss3
|
acyl-CoA synthetase short-chain family member 3 |
| chr5_+_7960445 | 1.09 |
ENSMUST00000115421.1
|
Steap4
|
STEAP family member 4 |
| chr5_-_105343929 | 1.07 |
ENSMUST00000183149.1
|
Gbp11
|
guanylate binding protein 11 |
| chr17_+_56628118 | 0.89 |
ENSMUST00000112979.2
|
Catsperd
|
catsper channel auxiliary subunit delta |
| chrM_+_7005 | 0.87 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr5_+_137981512 | 0.85 |
ENSMUST00000035390.5
|
Azgp1
|
alpha-2-glycoprotein 1, zinc |
| chr14_-_18894255 | 0.84 |
ENSMUST00000124353.1
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr7_-_127895578 | 0.81 |
ENSMUST00000033074.6
|
Vkorc1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr5_+_30921825 | 0.78 |
ENSMUST00000117435.1
|
Khk
|
ketohexokinase |
| chr11_-_69696058 | 0.77 |
ENSMUST00000180587.1
ENSMUST00000094070.4 |
Tnfsf12
BC096441
|
tumor necrosis factor (ligand) superfamily, member 12 cDNA sequence BC096441 |
| chr15_+_10981747 | 0.72 |
ENSMUST00000070877.5
|
Amacr
|
alpha-methylacyl-CoA racemase |
| chr16_+_23058250 | 0.69 |
ENSMUST00000039492.6
ENSMUST00000023589.8 ENSMUST00000089902.6 |
Kng1
|
kininogen 1 |
| chr9_-_108263706 | 0.66 |
ENSMUST00000171412.1
|
Dag1
|
dystroglycan 1 |
| chr15_+_10358525 | 0.66 |
ENSMUST00000110540.1
ENSMUST00000110541.1 ENSMUST00000110542.1 |
Agxt2
|
alanine-glyoxylate aminotransferase 2 |
| chr7_-_141434532 | 0.62 |
ENSMUST00000133021.1
ENSMUST00000106007.3 ENSMUST00000150026.1 ENSMUST00000133206.2 |
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr15_+_10358556 | 0.62 |
ENSMUST00000022858.7
|
Agxt2
|
alanine-glyoxylate aminotransferase 2 |
| chr3_-_116968969 | 0.60 |
ENSMUST00000143611.1
ENSMUST00000040097.7 |
Palmd
|
palmdelphin |
| chr11_+_105975204 | 0.57 |
ENSMUST00000001964.7
|
Ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr16_-_23029062 | 0.56 |
ENSMUST00000115349.2
|
Kng2
|
kininogen 2 |
| chr5_+_30921867 | 0.56 |
ENSMUST00000123885.1
|
Khk
|
ketohexokinase |
| chr16_-_23029080 | 0.55 |
ENSMUST00000100046.2
|
Kng2
|
kininogen 2 |
| chr18_+_37400845 | 0.55 |
ENSMUST00000057228.1
|
Pcdhb9
|
protocadherin beta 9 |
| chr9_+_85312773 | 0.54 |
ENSMUST00000113205.1
|
Gm11114
|
predicted gene 11114 |
| chr11_-_69695802 | 0.54 |
ENSMUST00000108649.1
ENSMUST00000174159.1 ENSMUST00000181810.1 |
BC096441
Tnfsf12
|
cDNA sequence BC096441 tumor necrosis factor (ligand) superfamily, member 12 |
| chr11_+_70166696 | 0.54 |
ENSMUST00000178567.1
|
Clec10a
|
C-type lectin domain family 10, member A |
| chr9_-_53610329 | 0.53 |
ENSMUST00000034547.5
|
Acat1
|
acetyl-Coenzyme A acetyltransferase 1 |
| chr2_+_147364989 | 0.53 |
ENSMUST00000109968.2
|
Pax1
|
paired box gene 1 |
| chrM_-_14060 | 0.51 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chrM_+_14138 | 0.51 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chrM_+_7759 | 0.50 |
ENSMUST00000082407.1
ENSMUST00000082408.1 |
mt-Atp8
mt-Atp6
|
mitochondrially encoded ATP synthase 8 mitochondrially encoded ATP synthase 6 |
| chr7_-_19665005 | 0.49 |
ENSMUST00000055242.9
|
Clptm1
|
cleft lip and palate associated transmembrane protein 1 |
| chrM_+_11734 | 0.49 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr19_-_28963863 | 0.49 |
ENSMUST00000161813.1
|
4430402I18Rik
|
RIKEN cDNA 4430402I18 gene |
| chr10_+_53337686 | 0.47 |
ENSMUST00000046221.6
ENSMUST00000163319.1 |
Pln
|
phospholamban |
| chr2_+_128591205 | 0.47 |
ENSMUST00000155430.1
|
Gm355
|
predicted gene 355 |
| chr17_-_59013264 | 0.46 |
ENSMUST00000174122.1
ENSMUST00000025065.5 |
Nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr7_-_126922887 | 0.45 |
ENSMUST00000134134.1
ENSMUST00000119781.1 ENSMUST00000121612.2 |
Tmem219
|
transmembrane protein 219 |
| chr15_+_101266839 | 0.45 |
ENSMUST00000023779.6
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr3_+_57425314 | 0.42 |
ENSMUST00000029377.7
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr5_+_43672289 | 0.40 |
ENSMUST00000156034.1
|
Cc2d2a
|
coiled-coil and C2 domain containing 2A |
| chr12_+_37242030 | 0.40 |
ENSMUST00000160390.1
|
Agmo
|
alkylglycerol monooxygenase |
| chr17_-_73950172 | 0.37 |
ENSMUST00000024866.4
|
Xdh
|
xanthine dehydrogenase |
| chr1_-_128359610 | 0.37 |
ENSMUST00000027601.4
|
Mcm6
|
minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) |
| chr11_+_70166623 | 0.37 |
ENSMUST00000102571.3
ENSMUST00000178945.1 ENSMUST00000000327.6 |
Clec10a
|
C-type lectin domain family 10, member A |
| chrY_-_38800793 | 0.36 |
ENSMUST00000180177.1
|
Gm20901
|
predicted gene, 20901 |
| chr18_+_37725706 | 0.35 |
ENSMUST00000066149.6
|
Pcdhga9
|
protocadherin gamma subfamily A, 9 |
| chr17_+_35236556 | 0.33 |
ENSMUST00000068261.8
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chrM_+_8600 | 0.32 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr3_-_89411781 | 0.31 |
ENSMUST00000107429.3
ENSMUST00000129308.2 ENSMUST00000107426.1 ENSMUST00000050398.4 ENSMUST00000162701.1 |
Flad1
|
RFad1, flavin adenine dinucleotide synthetase, homolog (yeast) |
| chr1_-_66863265 | 0.31 |
ENSMUST00000027153.5
|
Acadl
|
acyl-Coenzyme A dehydrogenase, long-chain |
| chr9_+_109054839 | 0.29 |
ENSMUST00000154184.1
|
Shisa5
|
shisa homolog 5 (Xenopus laevis) |
| chr6_+_134396316 | 0.27 |
ENSMUST00000032321.4
|
Bcl2l14
|
BCL2-like 14 (apoptosis facilitator) |
| chr16_-_8425104 | 0.27 |
ENSMUST00000023400.2
|
Cldn26
|
claudin 26 |
| chr6_+_40628824 | 0.27 |
ENSMUST00000071535.6
|
Mgam
|
maltase-glucoamylase |
| chr5_+_121660528 | 0.26 |
ENSMUST00000031414.8
|
Brap
|
BRCA1 associated protein |
| chr1_-_65186456 | 0.26 |
ENSMUST00000169032.1
|
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr10_+_79960145 | 0.26 |
ENSMUST00000045247.7
|
Wdr18
|
WD repeat domain 18 |
| chr13_+_49504774 | 0.25 |
ENSMUST00000051504.7
|
Ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr2_-_6951680 | 0.25 |
ENSMUST00000076071.2
|
Gm10115
|
predicted gene 10115 |
| chr7_+_29170345 | 0.25 |
ENSMUST00000033886.7
|
Ggn
|
gametogenetin |
| chr13_-_110280103 | 0.24 |
ENSMUST00000167824.1
|
Rab3c
|
RAB3C, member RAS oncogene family |
| chr14_-_70524068 | 0.24 |
ENSMUST00000022692.3
|
Sftpc
|
surfactant associated protein C |
| chr14_+_76476913 | 0.23 |
ENSMUST00000140251.2
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr17_-_35516780 | 0.23 |
ENSMUST00000160885.1
ENSMUST00000159009.1 ENSMUST00000161012.1 |
Tcf19
|
transcription factor 19 |
| chr17_-_56036546 | 0.23 |
ENSMUST00000003268.9
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr14_+_21481428 | 0.22 |
ENSMUST00000182996.1
|
Kat6b
|
K(lysine) acetyltransferase 6B |
| chr7_-_84409959 | 0.22 |
ENSMUST00000085077.3
|
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr10_+_57650990 | 0.22 |
ENSMUST00000175852.1
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr11_-_67052563 | 0.21 |
ENSMUST00000116363.1
|
Adprm
|
ADP-ribose/CDP-alcohol diphosphatase, manganese dependent |
| chr8_-_4216912 | 0.21 |
ENSMUST00000177491.1
|
BC068157
|
cDNA sequence BC068157 |
| chr2_-_9883993 | 0.21 |
ENSMUST00000114915.2
|
9230102O04Rik
|
RIKEN cDNA 9230102O04 gene |
| chr5_-_100373484 | 0.21 |
ENSMUST00000182433.1
|
Sec31a
|
Sec31 homolog A (S. cerevisiae) |
| chr9_+_109816618 | 0.20 |
ENSMUST00000118732.1
|
Spink8
|
serine peptidase inhibitor, Kazal type 8 |
| chr6_-_3399545 | 0.19 |
ENSMUST00000120087.3
|
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr1_-_90843916 | 0.19 |
ENSMUST00000130846.2
ENSMUST00000097653.4 ENSMUST00000056925.9 |
Col6a3
|
collagen, type VI, alpha 3 |
| chr5_+_140735526 | 0.19 |
ENSMUST00000120630.2
|
Amz1
|
archaelysin family metallopeptidase 1 |
| chr8_-_109962127 | 0.18 |
ENSMUST00000001722.7
ENSMUST00000051430.6 |
Marveld3
|
MARVEL (membrane-associating) domain containing 3 |
| chr5_-_108795352 | 0.17 |
ENSMUST00000004943.1
|
Tmed11
|
transmembrane emp24 protein transport domain containing |
| chr10_+_3872667 | 0.17 |
ENSMUST00000136671.1
ENSMUST00000042438.6 |
Plekhg1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr5_-_50058908 | 0.16 |
ENSMUST00000030971.5
|
Gpr125
|
G protein-coupled receptor 125 |
| chr9_+_13662460 | 0.16 |
ENSMUST00000177755.1
|
Maml2
|
mastermind like 2 (Drosophila) |
| chr8_+_111643425 | 0.15 |
ENSMUST00000077791.6
|
Zfp1
|
zinc finger protein 1 |
| chr11_-_99521336 | 0.15 |
ENSMUST00000107445.1
|
Krt39
|
keratin 39 |
| chr17_-_17883920 | 0.15 |
ENSMUST00000061516.7
|
Fpr1
|
formyl peptide receptor 1 |
| chr10_+_57650977 | 0.14 |
ENSMUST00000066028.6
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr6_-_91515878 | 0.14 |
ENSMUST00000032182.3
|
Xpc
|
xeroderma pigmentosum, complementation group C |
| chr17_+_22689771 | 0.13 |
ENSMUST00000055305.8
|
Gm9805
|
predicted gene 9805 |
| chr7_-_121981669 | 0.13 |
ENSMUST00000057576.7
|
Cog7
|
component of oligomeric golgi complex 7 |
| chr3_-_92827239 | 0.12 |
ENSMUST00000072363.4
|
Kprp
|
keratinocyte expressed, proline-rich |
| chr5_-_113310697 | 0.12 |
ENSMUST00000154248.1
|
Sgsm1
|
small G protein signaling modulator 1 |
| chr17_-_42692244 | 0.12 |
ENSMUST00000170723.1
ENSMUST00000164524.1 ENSMUST00000024711.4 ENSMUST00000167993.1 |
Gpr115
|
G protein-coupled receptor 115 |
| chr10_-_130280218 | 0.12 |
ENSMUST00000061571.3
|
Neurod4
|
neurogenic differentiation 4 |
| chr11_+_67052641 | 0.12 |
ENSMUST00000108690.3
ENSMUST00000092996.4 |
Sco1
|
SCO cytochrome oxidase deficient homolog 1 (yeast) |
| chr8_-_54529951 | 0.11 |
ENSMUST00000067476.8
|
Spcs3
|
signal peptidase complex subunit 3 homolog (S. cerevisiae) |
| chr7_+_45896941 | 0.10 |
ENSMUST00000069772.7
ENSMUST00000107716.1 |
Tmem143
|
transmembrane protein 143 |
| chr10_-_81014902 | 0.10 |
ENSMUST00000126317.1
ENSMUST00000092285.3 ENSMUST00000117805.1 |
Gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr3_-_33082004 | 0.10 |
ENSMUST00000108225.3
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr2_-_92540967 | 0.10 |
ENSMUST00000142249.2
|
Gm13817
|
predicted gene 13817 |
| chr5_+_114896936 | 0.10 |
ENSMUST00000031542.9
ENSMUST00000146072.1 ENSMUST00000150361.1 |
Oasl2
|
2'-5' oligoadenylate synthetase-like 2 |
| chr8_-_19114830 | 0.10 |
ENSMUST00000062113.7
|
Defb12
|
defensin beta 12 |
| chr14_-_50681645 | 0.09 |
ENSMUST00000078075.4
|
Olfr747
|
olfactory receptor 747 |
| chr12_+_87200524 | 0.09 |
ENSMUST00000182869.1
|
Samd15
|
sterile alpha motif domain containing 15 |
| chr11_-_96111962 | 0.08 |
ENSMUST00000097162.5
ENSMUST00000068686.6 |
Calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr1_+_44119952 | 0.08 |
ENSMUST00000114709.2
|
Bivm
|
basic, immunoglobulin-like variable motif containing |
| chr11_-_115627948 | 0.08 |
ENSMUST00000154623.1
ENSMUST00000106503.3 ENSMUST00000141614.1 |
Slc25a19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr9_+_109816571 | 0.07 |
ENSMUST00000055821.5
|
Spink8
|
serine peptidase inhibitor, Kazal type 8 |
| chr16_+_44820728 | 0.07 |
ENSMUST00000114626.3
|
Cd200r4
|
CD200 receptor 4 |
| chr12_+_36157124 | 0.07 |
ENSMUST00000041640.3
|
Ankmy2
|
ankyrin repeat and MYND domain containing 2 |
| chr14_-_63037833 | 0.07 |
ENSMUST00000111207.1
ENSMUST00000100490.2 |
Defb30
|
defensin beta 30 |
| chr3_+_94443315 | 0.07 |
ENSMUST00000029786.7
ENSMUST00000098876.3 |
Mrpl9
|
mitochondrial ribosomal protein L9 |
| chr16_+_34651188 | 0.07 |
ENSMUST00000023530.4
|
Ropn1
|
ropporin, rhophilin associated protein 1 |
| chr2_-_94010807 | 0.07 |
ENSMUST00000111240.1
|
Alkbh3
|
alkB, alkylation repair homolog 3 (E. coli) |
| chr15_+_22549022 | 0.07 |
ENSMUST00000163361.1
|
Cdh18
|
cadherin 18 |
| chrY_-_79149787 | 0.07 |
ENSMUST00000179922.1
|
Gm20917
|
predicted gene, 20917 |
| chr10_+_37139558 | 0.06 |
ENSMUST00000062667.3
|
5930403N24Rik
|
RIKEN cDNA 5930403N24 gene |
| chr7_+_45897429 | 0.06 |
ENSMUST00000140243.1
|
Tmem143
|
transmembrane protein 143 |
| chr6_-_148896150 | 0.06 |
ENSMUST00000072324.5
ENSMUST00000111569.2 |
Caprin2
|
caprin family member 2 |
| chr14_+_51114455 | 0.06 |
ENSMUST00000100645.3
|
Eddm3b
|
epididymal protein 3B |
| chrX_-_92016183 | 0.05 |
ENSMUST00000099471.2
ENSMUST00000072269.1 |
Mageb1
|
melanoma antigen, family B, 1 |
| chr3_+_83026147 | 0.05 |
ENSMUST00000166581.1
ENSMUST00000029630.9 |
Fga
|
fibrinogen alpha chain |
| chr7_+_28863831 | 0.05 |
ENSMUST00000138272.1
|
Lgals7
|
lectin, galactose binding, soluble 7 |
| chr18_+_77375065 | 0.04 |
ENSMUST00000123410.1
|
Loxhd1
|
lipoxygenase homology domains 1 |
| chr7_-_104164812 | 0.04 |
ENSMUST00000154555.1
ENSMUST00000051137.8 |
E030002O03Rik
|
RIKEN cDNA E030002O03 gene |
| chr8_-_4217133 | 0.03 |
ENSMUST00000175906.1
|
BC068157
|
cDNA sequence BC068157 |
| chr1_+_21349598 | 0.03 |
ENSMUST00000088407.3
|
Khdc1a
|
KH domain containing 1A |
| chr9_-_110989611 | 0.03 |
ENSMUST00000084922.5
|
Rtp3
|
receptor transporter protein 3 |
| chrX_-_7041619 | 0.03 |
ENSMUST00000115752.1
|
Ccnb3
|
cyclin B3 |
| chr12_+_10390756 | 0.03 |
ENSMUST00000020947.5
|
Rdh14
|
retinol dehydrogenase 14 (all-trans and 9-cis) |
| chr8_-_15046047 | 0.02 |
ENSMUST00000050493.3
ENSMUST00000123331.1 |
BB014433
|
expressed sequence BB014433 |
| chr19_+_46707443 | 0.02 |
ENSMUST00000003655.7
|
As3mt
|
arsenic (+3 oxidation state) methyltransferase |
| chrY_-_88953082 | 0.02 |
ENSMUST00000179463.1
|
Gm21720
|
predicted gene, 21720 |
| chr19_+_34217588 | 0.02 |
ENSMUST00000119603.1
|
Stambpl1
|
STAM binding protein like 1 |
| chr1_+_189728264 | 0.02 |
ENSMUST00000097442.2
|
Ptpn14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr7_+_11005722 | 0.01 |
ENSMUST00000131379.2
|
Zscan4c
|
zinc finger and SCAN domain containing 4C |
| chr6_-_40900368 | 0.01 |
ENSMUST00000063523.3
|
Prss58
|
protease, serine 58 |
| chr15_-_98165560 | 0.01 |
ENSMUST00000123922.1
|
Asb8
|
ankyrin repeat and SOCS box-containing 8 |
| chr14_-_79223876 | 0.01 |
ENSMUST00000040802.4
|
Zfp957
|
zinc finger protein 957 |
| chr16_-_91618986 | 0.01 |
ENSMUST00000143058.1
ENSMUST00000049244.8 ENSMUST00000169982.1 ENSMUST00000133731.1 |
Dnajc28
|
DnaJ (Hsp40) homolog, subfamily C, member 28 |
| chr15_-_98165613 | 0.00 |
ENSMUST00000143400.1
|
Asb8
|
ankyrin repeat and SOCS box-containing 8 |
| chr3_-_95217877 | 0.00 |
ENSMUST00000136139.1
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr12_+_4843303 | 0.00 |
ENSMUST00000053034.4
|
BC068281
|
cDNA sequence BC068281 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Aire
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0042853 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) L-alanine catabolic process(GO:0042853) |
| 0.4 | 1.5 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.3 | 1.3 | GO:0061625 | fructose catabolic process(GO:0006001) response to sucrose(GO:0009744) response to disaccharide(GO:0034285) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.3 | 0.8 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.2 | 0.6 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.2 | 0.5 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.2 | 0.7 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.1 | 0.9 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) |
| 0.1 | 1.5 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.5 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.1 | 1.1 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 0.8 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.3 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.1 | 1.5 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.1 | 0.3 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.5 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.5 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 2.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.5 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.4 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.1 | 0.7 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.1 | 0.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.5 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 1.9 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 1.0 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.3 | GO:1902572 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 1.4 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.6 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 1.6 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.1 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.0 | 1.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.1 | GO:0035553 | oxidative single-stranded DNA demethylation(GO:0035552) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.2 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.5 | GO:2000398 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 0.7 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.1 | 2.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.5 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.0 | 1.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 4.4 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.0 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.1 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.9 | GO:0072562 | blood microparticle(GO:0072562) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 0.4 | 2.6 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.4 | 1.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.2 | 1.2 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.2 | 1.3 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.1 | 0.6 | GO:0031711 | tripeptidyl-peptidase activity(GO:0008240) bradykinin receptor binding(GO:0031711) metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.8 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 1.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.4 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.1 | 0.5 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.1 | 0.3 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 3.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.5 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.3 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.1 | 1.5 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.4 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.1 | 0.7 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.5 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 1.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.3 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.6 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.8 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 1.2 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.0 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.7 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 1.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0051747 | cytosine C-5 DNA demethylase activity(GO:0051747) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.4 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 1.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 1.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 0.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 1.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |