Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
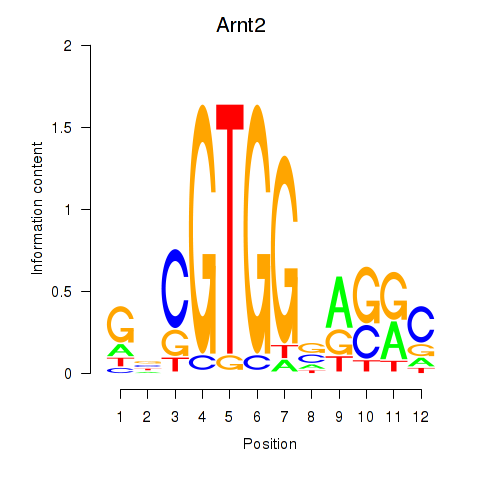
Results for Arnt2
Z-value: 0.26
Transcription factors associated with Arnt2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arnt2
|
ENSMUSG00000015709.8 | aryl hydrocarbon receptor nuclear translocator 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Arnt2 | mm10_v2_chr7_-_84409959_84410002 | 0.74 | 2.8e-07 | Click! |
Activity profile of Arnt2 motif
Sorted Z-values of Arnt2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_54085542 | 0.75 |
ENSMUST00000100089.2
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr2_+_79255500 | 0.59 |
ENSMUST00000099972.4
|
Itga4
|
integrin alpha 4 |
| chr1_-_58973421 | 0.57 |
ENSMUST00000173590.1
ENSMUST00000027186.5 |
Trak2
|
trafficking protein, kinesin binding 2 |
| chr7_+_130865835 | 0.45 |
ENSMUST00000075181.4
ENSMUST00000048180.5 |
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr2_-_91649785 | 0.44 |
ENSMUST00000111333.1
|
Zfp408
|
zinc finger protein 408 |
| chr2_+_27886416 | 0.43 |
ENSMUST00000028280.7
|
Col5a1
|
collagen, type V, alpha 1 |
| chr11_+_43528759 | 0.42 |
ENSMUST00000050574.6
|
Ccnjl
|
cyclin J-like |
| chr7_+_130865756 | 0.39 |
ENSMUST00000120441.1
|
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr5_-_115300957 | 0.38 |
ENSMUST00000009157.3
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr5_-_115300912 | 0.37 |
ENSMUST00000112090.1
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr2_-_91649751 | 0.37 |
ENSMUST00000099714.3
|
Zfp408
|
zinc finger protein 408 |
| chr4_-_123116686 | 0.34 |
ENSMUST00000166337.1
|
Gm17244
|
predicted gene, 17244 |
| chr7_-_118491912 | 0.33 |
ENSMUST00000178344.1
|
Itpripl2
|
inositol 1,4,5-triphosphate receptor interacting protein-like 2 |
| chr2_+_14604252 | 0.32 |
ENSMUST00000114723.2
|
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr5_-_135251209 | 0.32 |
ENSMUST00000062572.2
|
Fzd9
|
frizzled homolog 9 (Drosophila) |
| chr7_-_45091713 | 0.31 |
ENSMUST00000141576.1
|
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr4_-_41275091 | 0.30 |
ENSMUST00000030143.6
ENSMUST00000108068.1 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr4_+_123116246 | 0.29 |
ENSMUST00000102648.4
|
Oxct2b
|
3-oxoacid CoA transferase 2B |
| chr4_-_123323603 | 0.25 |
ENSMUST00000102640.1
|
Oxct2a
|
3-oxoacid CoA transferase 2A |
| chr19_+_43612299 | 0.24 |
ENSMUST00000057178.9
|
Nkx2-3
|
NK2 homeobox 3 |
| chr19_-_5098418 | 0.22 |
ENSMUST00000025805.6
|
Cnih2
|
cornichon homolog 2 (Drosophila) |
| chr7_-_45092130 | 0.19 |
ENSMUST00000148175.1
|
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr18_+_31634368 | 0.19 |
ENSMUST00000178164.1
|
Sap130
|
Sin3A associated protein |
| chr13_+_13590402 | 0.19 |
ENSMUST00000110559.1
|
Lyst
|
lysosomal trafficking regulator |
| chr4_+_101496648 | 0.19 |
ENSMUST00000106930.1
ENSMUST00000154120.1 |
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr18_+_31634374 | 0.17 |
ENSMUST00000025109.7
|
Sap130
|
Sin3A associated protein |
| chr3_+_96697076 | 0.17 |
ENSMUST00000162778.2
ENSMUST00000064900.9 |
Pias3
|
protein inhibitor of activated STAT 3 |
| chrX_-_53269786 | 0.14 |
ENSMUST00000114841.1
ENSMUST00000071023.5 |
Fam122b
|
family with sequence similarity 122, member B |
| chr2_+_5951440 | 0.13 |
ENSMUST00000060092.6
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr7_-_45370559 | 0.11 |
ENSMUST00000003971.7
|
Lin7b
|
lin-7 homolog B (C. elegans) |
| chr8_+_83900706 | 0.11 |
ENSMUST00000045393.8
ENSMUST00000132500.1 ENSMUST00000152978.1 |
Lphn1
|
latrophilin 1 |
| chr3_+_96697100 | 0.10 |
ENSMUST00000107077.3
|
Pias3
|
protein inhibitor of activated STAT 3 |
| chr13_+_118714678 | 0.10 |
ENSMUST00000022246.8
|
Fgf10
|
fibroblast growth factor 10 |
| chr1_+_87327008 | 0.09 |
ENSMUST00000172794.1
ENSMUST00000164992.2 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr11_+_107547925 | 0.09 |
ENSMUST00000100305.1
ENSMUST00000075012.1 ENSMUST00000106746.1 |
Helz
|
helicase with zinc finger domain |
| chr1_+_87327044 | 0.09 |
ENSMUST00000173173.1
|
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr5_+_64803513 | 0.08 |
ENSMUST00000165536.1
|
Klf3
|
Kruppel-like factor 3 (basic) |
| chr1_+_59912972 | 0.04 |
ENSMUST00000036540.5
|
Fam117b
|
family with sequence similarity 117, member B |
| chr6_+_4601124 | 0.03 |
ENSMUST00000181734.1
ENSMUST00000141359.1 |
Casd1
|
CAS1 domain containing 1 |
| chr5_-_44226601 | 0.03 |
ENSMUST00000055128.7
|
Tapt1
|
transmembrane anterior posterior transformation 1 |
| chr1_+_87326997 | 0.03 |
ENSMUST00000027475.8
|
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr7_-_44869788 | 0.02 |
ENSMUST00000046575.9
|
Ptov1
|
prostate tumor over expressed gene 1 |
| chr1_-_52817503 | 0.02 |
ENSMUST00000162576.1
|
Inpp1
|
inositol polyphosphate-1-phosphatase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Arnt2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0050904 | diapedesis(GO:0050904) |
| 0.1 | 0.5 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.3 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 0.3 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.2 | GO:1901608 | regulation of vesicle transport along microtubule(GO:1901608) |
| 0.1 | 0.8 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.6 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.0 | 0.8 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.2 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.1 | GO:0061033 | bronchiole development(GO:0060435) secretion by lung epithelial cell involved in lung growth(GO:0061033) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.0 | 0.2 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.6 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 0.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.8 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |