Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
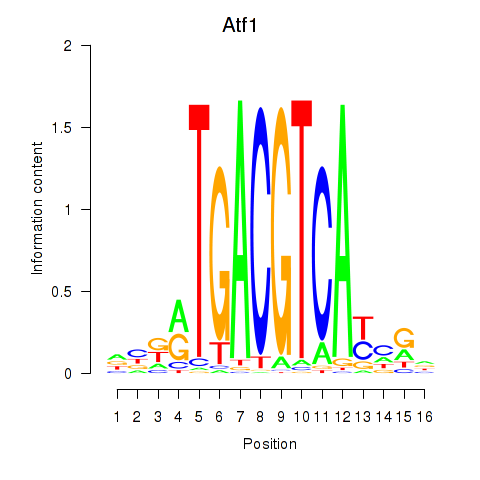
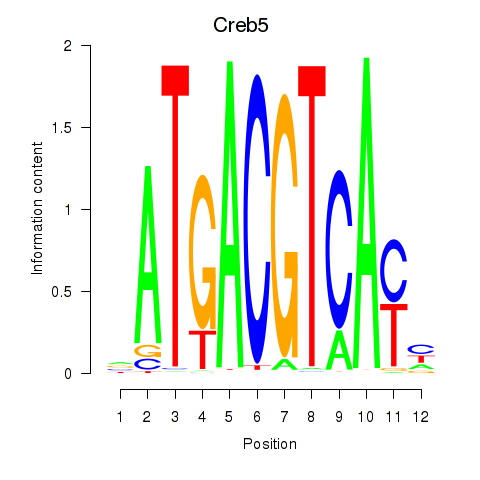
Results for Atf1_Creb5
Z-value: 0.76
Transcription factors associated with Atf1_Creb5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf1
|
ENSMUSG00000023027.6 | activating transcription factor 1 |
|
Creb5
|
ENSMUSG00000053007.6 | cAMP responsive element binding protein 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf1 | mm10_v2_chr15_+_100227819_100227856 | 0.44 | 7.2e-03 | Click! |
| Creb5 | mm10_v2_chr6_+_53573364_53573394 | -0.31 | 6.4e-02 | Click! |
Activity profile of Atf1_Creb5 motif
Sorted Z-values of Atf1_Creb5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_75571522 | 5.14 |
ENSMUST00000143226.1
ENSMUST00000124259.1 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr3_+_135212557 | 3.95 |
ENSMUST00000062893.7
|
Cenpe
|
centromere protein E |
| chr7_-_142095266 | 3.69 |
ENSMUST00000039926.3
|
Dusp8
|
dual specificity phosphatase 8 |
| chr6_-_86669136 | 3.40 |
ENSMUST00000001184.7
|
Mxd1
|
MAX dimerization protein 1 |
| chr2_+_109280738 | 3.17 |
ENSMUST00000028527.7
|
Kif18a
|
kinesin family member 18A |
| chr14_-_87141206 | 3.09 |
ENSMUST00000022599.7
|
Diap3
|
diaphanous homolog 3 (Drosophila) |
| chr8_-_92355764 | 3.09 |
ENSMUST00000180102.1
ENSMUST00000179421.1 ENSMUST00000179222.1 ENSMUST00000179029.1 |
4933436C20Rik
|
RIKEN cDNA 4933436C20 gene |
| chr14_-_87141114 | 2.99 |
ENSMUST00000168889.1
|
Diap3
|
diaphanous homolog 3 (Drosophila) |
| chr12_+_4082574 | 2.92 |
ENSMUST00000020986.7
|
Dnajc27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr8_-_85365341 | 2.66 |
ENSMUST00000121972.1
|
Mylk3
|
myosin light chain kinase 3 |
| chr8_-_85365317 | 2.64 |
ENSMUST00000034133.7
|
Mylk3
|
myosin light chain kinase 3 |
| chr3_+_40800054 | 2.62 |
ENSMUST00000168287.1
|
Plk4
|
polo-like kinase 4 |
| chr15_+_79347534 | 2.55 |
ENSMUST00000096350.3
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr8_+_23139064 | 2.54 |
ENSMUST00000033947.8
|
Ank1
|
ankyrin 1, erythroid |
| chr5_+_143548700 | 2.46 |
ENSMUST00000169329.1
ENSMUST00000067145.5 ENSMUST00000119488.1 ENSMUST00000118121.1 |
Fam220a
Fam220a
|
family with sequence similarity 220, member A family with sequence similarity 220, member A |
| chr8_+_23139030 | 2.43 |
ENSMUST00000121075.1
|
Ank1
|
ankyrin 1, erythroid |
| chr5_+_93206518 | 2.43 |
ENSMUST00000031330.4
|
2010109A12Rik
|
RIKEN cDNA 2010109A12 gene |
| chr7_+_44468020 | 2.35 |
ENSMUST00000117324.1
ENSMUST00000120852.1 ENSMUST00000118628.1 |
Josd2
|
Josephin domain containing 2 |
| chr3_+_40800013 | 2.31 |
ENSMUST00000026858.5
ENSMUST00000170825.1 |
Plk4
|
polo-like kinase 4 |
| chr8_+_116921735 | 2.30 |
ENSMUST00000034205.4
|
Cenpn
|
centromere protein N |
| chr3_+_127553462 | 2.21 |
ENSMUST00000043108.4
|
4930422G04Rik
|
RIKEN cDNA 4930422G04 gene |
| chr14_-_69503316 | 2.20 |
ENSMUST00000179116.2
|
Gm21464
|
predicted gene, 21464 |
| chr3_+_153844209 | 2.19 |
ENSMUST00000044089.3
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chr12_+_4082596 | 2.19 |
ENSMUST00000049584.5
|
Dnajc27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr7_+_44467980 | 2.12 |
ENSMUST00000035844.4
|
Josd2
|
Josephin domain containing 2 |
| chr7_+_44468051 | 2.11 |
ENSMUST00000118493.1
|
Josd2
|
Josephin domain containing 2 |
| chr6_+_113531675 | 2.04 |
ENSMUST00000036340.5
ENSMUST00000101051.2 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr1_+_174158722 | 2.03 |
ENSMUST00000068403.3
|
Olfr420
|
olfactory receptor 420 |
| chr14_-_69284982 | 2.02 |
ENSMUST00000183882.1
ENSMUST00000037064.4 |
Slc25a37
|
solute carrier family 25, member 37 |
| chr7_-_45526146 | 2.02 |
ENSMUST00000167273.1
ENSMUST00000042105.8 |
Ppp1r15a
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A |
| chr2_-_164745916 | 2.02 |
ENSMUST00000109328.1
ENSMUST00000043448.1 |
Wfdc3
Wfdc3
|
WAP four-disulfide core domain 3 WAP four-disulfide core domain 3 |
| chr3_-_36571952 | 1.98 |
ENSMUST00000029270.3
|
Ccna2
|
cyclin A2 |
| chr8_-_92356103 | 1.98 |
ENSMUST00000034183.3
|
4933436C20Rik
|
RIKEN cDNA 4933436C20 gene |
| chr14_-_69503220 | 1.95 |
ENSMUST00000180059.2
|
Gm21464
|
predicted gene, 21464 |
| chr6_-_83033422 | 1.88 |
ENSMUST00000089651.5
|
Dok1
|
docking protein 1 |
| chr6_-_129533267 | 1.87 |
ENSMUST00000181594.1
|
1700101I11Rik
|
RIKEN cDNA 1700101I11 gene |
| chrX_+_9283764 | 1.83 |
ENSMUST00000177926.1
|
1700012L04Rik
|
RIKEN cDNA 1700012L04 gene |
| chr10_-_30200492 | 1.82 |
ENSMUST00000099985.4
|
Cenpw
|
centromere protein W |
| chr14_+_47472628 | 1.82 |
ENSMUST00000095941.2
|
Fbxo34
|
F-box protein 34 |
| chrX_-_8145713 | 1.77 |
ENSMUST00000115615.2
ENSMUST00000115616.1 ENSMUST00000115621.2 |
Rbm3
|
RNA binding motif protein 3 |
| chr7_-_30664986 | 1.76 |
ENSMUST00000019697.8
|
Haus5
|
HAUS augmin-like complex, subunit 5 |
| chr13_+_22043189 | 1.69 |
ENSMUST00000110452.1
|
Hist1h2bj
|
histone cluster 1, H2bj |
| chr8_-_84840627 | 1.68 |
ENSMUST00000003911.6
ENSMUST00000109761.2 ENSMUST00000128035.1 |
Rad23a
|
RAD23a homolog (S. cerevisiae) |
| chr11_+_113619318 | 1.67 |
ENSMUST00000146390.2
ENSMUST00000106630.1 |
Sstr2
|
somatostatin receptor 2 |
| chr4_+_130360132 | 1.65 |
ENSMUST00000105994.3
|
Snrnp40
|
small nuclear ribonucleoprotein 40 (U5) |
| chr7_-_133702515 | 1.63 |
ENSMUST00000153698.1
|
Uros
|
uroporphyrinogen III synthase |
| chrX_-_8145679 | 1.61 |
ENSMUST00000115619.1
ENSMUST00000115617.3 ENSMUST00000040010.3 |
Rbm3
|
RNA binding motif protein 3 |
| chr12_-_69159109 | 1.60 |
ENSMUST00000037023.8
|
Rps29
|
ribosomal protein S29 |
| chr13_+_75707484 | 1.59 |
ENSMUST00000001583.6
|
Ell2
|
elongation factor RNA polymerase II 2 |
| chr13_-_22042949 | 1.58 |
ENSMUST00000091741.4
|
Hist1h2ag
|
histone cluster 1, H2ag |
| chr2_+_148798785 | 1.58 |
ENSMUST00000028931.3
ENSMUST00000109947.1 |
Cst8
|
cystatin 8 (cystatin-related epididymal spermatogenic) |
| chr18_-_67549173 | 1.55 |
ENSMUST00000115050.1
|
Spire1
|
spire homolog 1 (Drosophila) |
| chr3_-_103791075 | 1.54 |
ENSMUST00000106845.2
ENSMUST00000029438.8 ENSMUST00000121324.1 |
Hipk1
|
homeodomain interacting protein kinase 1 |
| chr1_+_134455524 | 1.51 |
ENSMUST00000112232.1
ENSMUST00000027725.4 ENSMUST00000116528.1 |
Klhl12
|
kelch-like 12 |
| chr5_+_123076275 | 1.49 |
ENSMUST00000067505.8
ENSMUST00000111619.3 ENSMUST00000160344.1 |
Tmem120b
|
transmembrane protein 120B |
| chrX_-_88760312 | 1.47 |
ENSMUST00000182943.1
|
Gm27000
|
predicted gene, 27000 |
| chr11_-_17008647 | 1.46 |
ENSMUST00000102881.3
|
Plek
|
pleckstrin |
| chr9_+_106514572 | 1.45 |
ENSMUST00000085113.3
|
Iqcf5
|
IQ motif containing F5 |
| chr3_-_103791537 | 1.44 |
ENSMUST00000118317.1
|
Hipk1
|
homeodomain interacting protein kinase 1 |
| chr14_+_47472547 | 1.43 |
ENSMUST00000168833.1
ENSMUST00000163324.1 ENSMUST00000043112.7 |
Fbxo34
|
F-box protein 34 |
| chr17_-_71526819 | 1.42 |
ENSMUST00000024851.9
|
Ndc80
|
NDC80 homolog, kinetochore complex component (S. cerevisiae) |
| chr6_-_131293187 | 1.36 |
ENSMUST00000032307.5
|
Magohb
|
mago-nashi homolog B (Drosophila) |
| chr2_-_132253227 | 1.35 |
ENSMUST00000028817.6
|
Pcna
|
proliferating cell nuclear antigen |
| chr11_+_69965396 | 1.35 |
ENSMUST00000018713.6
|
Cldn7
|
claudin 7 |
| chr11_-_3504766 | 1.30 |
ENSMUST00000044507.5
|
Inpp5j
|
inositol polyphosphate 5-phosphatase J |
| chr15_+_99074968 | 1.29 |
ENSMUST00000039665.6
|
Troap
|
trophinin associated protein |
| chr12_+_73286868 | 1.29 |
ENSMUST00000153941.1
ENSMUST00000122920.1 ENSMUST00000101313.3 |
Slc38a6
|
solute carrier family 38, member 6 |
| chr8_-_123949201 | 1.25 |
ENSMUST00000044795.7
|
Nup133
|
nucleoporin 133 |
| chr4_-_107810948 | 1.23 |
ENSMUST00000097930.1
|
B230314M03Rik
|
RIKEN cDNA B230314M03 gene |
| chr19_-_24961545 | 1.23 |
ENSMUST00000025815.8
|
Cbwd1
|
COBW domain containing 1 |
| chr11_-_59163281 | 1.21 |
ENSMUST00000069631.2
|
Iba57
|
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) |
| chr10_-_111997204 | 1.20 |
ENSMUST00000074805.5
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr8_+_23139157 | 1.18 |
ENSMUST00000174435.1
|
Ank1
|
ankyrin 1, erythroid |
| chr5_+_101765120 | 1.16 |
ENSMUST00000031273.8
|
Cds1
|
CDP-diacylglycerol synthase 1 |
| chr7_+_141061274 | 1.16 |
ENSMUST00000048002.5
|
B4galnt4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4 |
| chr19_+_6105774 | 1.14 |
ENSMUST00000044451.3
|
Naaladl1
|
N-acetylated alpha-linked acidic dipeptidase-like 1 |
| chr2_+_120609383 | 1.13 |
ENSMUST00000124187.1
|
Haus2
|
HAUS augmin-like complex, subunit 2 |
| chr19_+_37376359 | 1.10 |
ENSMUST00000012587.3
|
Kif11
|
kinesin family member 11 |
| chr12_+_109540979 | 1.09 |
ENSMUST00000129245.1
ENSMUST00000143836.1 ENSMUST00000124106.1 |
Meg3
|
maternally expressed 3 |
| chr6_-_124464772 | 1.09 |
ENSMUST00000008297.4
|
Clstn3
|
calsyntenin 3 |
| chr7_+_125603420 | 1.09 |
ENSMUST00000033000.6
|
Il21r
|
interleukin 21 receptor |
| chr4_-_134245579 | 1.08 |
ENSMUST00000030644.7
|
Zfp593
|
zinc finger protein 593 |
| chr1_-_134079114 | 1.07 |
ENSMUST00000020692.6
|
Btg2
|
B cell translocation gene 2, anti-proliferative |
| chr13_+_19623163 | 1.07 |
ENSMUST00000002883.5
|
Sfrp4
|
secreted frizzled-related protein 4 |
| chr5_+_31698050 | 1.06 |
ENSMUST00000114507.3
ENSMUST00000063813.4 ENSMUST00000071531.5 ENSMUST00000131995.1 |
Bre
|
brain and reproductive organ-expressed protein |
| chr4_-_131838231 | 1.04 |
ENSMUST00000030741.2
ENSMUST00000105987.2 |
Ptpru
|
protein tyrosine phosphatase, receptor type, U |
| chr6_+_30723541 | 1.03 |
ENSMUST00000115127.1
|
Mest
|
mesoderm specific transcript |
| chr7_+_3703979 | 1.02 |
ENSMUST00000006496.8
|
Rps9
|
ribosomal protein S9 |
| chr7_+_45621805 | 1.00 |
ENSMUST00000033100.4
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr2_+_30834972 | 1.00 |
ENSMUST00000113592.2
|
Prrx2
|
paired related homeobox 2 |
| chr5_+_24164963 | 0.99 |
ENSMUST00000049887.6
|
Nupl2
|
nucleoporin like 2 |
| chr7_+_3704307 | 0.99 |
ENSMUST00000108624.1
ENSMUST00000126562.1 |
Rps9
|
ribosomal protein S9 |
| chr11_-_97782377 | 0.98 |
ENSMUST00000128801.1
|
Rpl23
|
ribosomal protein L23 |
| chr2_+_164746028 | 0.97 |
ENSMUST00000109327.3
|
Dnttip1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr11_-_88718078 | 0.97 |
ENSMUST00000092794.5
|
Msi2
|
musashi RNA-binding protein 2 |
| chr7_+_3704025 | 0.97 |
ENSMUST00000108623.1
ENSMUST00000139818.1 ENSMUST00000108625.1 |
Rps9
|
ribosomal protein S9 |
| chr19_+_59260878 | 0.97 |
ENSMUST00000026084.3
|
Slc18a2
|
solute carrier family 18 (vesicular monoamine), member 2 |
| chr7_+_30413744 | 0.96 |
ENSMUST00000032800.9
|
Tyrobp
|
TYRO protein tyrosine kinase binding protein |
| chr19_-_61228396 | 0.96 |
ENSMUST00000076046.6
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr13_+_21945084 | 0.95 |
ENSMUST00000176511.1
ENSMUST00000102978.1 ENSMUST00000152258.2 |
Zfp184
|
zinc finger protein 184 (Kruppel-like) |
| chr13_+_5861489 | 0.95 |
ENSMUST00000000080.6
|
Klf6
|
Kruppel-like factor 6 |
| chr6_+_83034173 | 0.93 |
ENSMUST00000000707.2
ENSMUST00000101257.3 |
Loxl3
|
lysyl oxidase-like 3 |
| chr9_+_53850243 | 0.93 |
ENSMUST00000048485.5
|
Sln
|
sarcolipin |
| chr2_+_172472512 | 0.92 |
ENSMUST00000029007.2
|
Fam209
|
family with sequence similarity 209 |
| chr4_-_132075250 | 0.91 |
ENSMUST00000105970.1
ENSMUST00000105975.1 |
Epb4.1
|
erythrocyte protein band 4.1 |
| chr4_+_41348996 | 0.91 |
ENSMUST00000072866.5
ENSMUST00000108060.3 |
Ubap1
|
ubiquitin-associated protein 1 |
| chr2_+_164745979 | 0.90 |
ENSMUST00000017443.7
ENSMUST00000109326.3 |
Dnttip1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr2_-_23155864 | 0.90 |
ENSMUST00000028119.6
|
Mastl
|
microtubule associated serine/threonine kinase-like |
| chr11_+_59208321 | 0.90 |
ENSMUST00000020719.6
|
2310033P09Rik
|
RIKEN cDNA 2310033P09 gene |
| chr16_+_38346986 | 0.89 |
ENSMUST00000050273.8
ENSMUST00000120495.1 ENSMUST00000119704.1 |
Cox17
Gm21987
|
cytochrome c oxidase assembly protein 17 predicted gene 21987 |
| chr5_-_137149320 | 0.89 |
ENSMUST00000041226.8
|
Muc3
|
mucin 3, intestinal |
| chr17_-_31512253 | 0.88 |
ENSMUST00000166626.1
|
Wdr4
|
WD repeat domain 4 |
| chr4_+_129461581 | 0.88 |
ENSMUST00000048162.8
ENSMUST00000138013.1 |
Bsdc1
|
BSD domain containing 1 |
| chr11_-_88718223 | 0.88 |
ENSMUST00000107909.1
|
Msi2
|
musashi RNA-binding protein 2 |
| chr1_+_107511416 | 0.88 |
ENSMUST00000009356.4
|
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| chr7_-_83884289 | 0.88 |
ENSMUST00000094216.3
|
Mesdc1
|
mesoderm development candidate 1 |
| chr5_+_33820695 | 0.87 |
ENSMUST00000075812.4
ENSMUST00000114397.2 ENSMUST00000155880.1 |
Whsc1
|
Wolf-Hirschhorn syndrome candidate 1 (human) |
| chr1_+_107511489 | 0.87 |
ENSMUST00000064916.2
|
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| chr19_-_4625612 | 0.87 |
ENSMUST00000025823.3
|
Rce1
|
RCE1 homolog, prenyl protein peptidase (S. cerevisiae) |
| chr3_-_127553233 | 0.85 |
ENSMUST00000029588.5
|
Larp7
|
La ribonucleoprotein domain family, member 7 |
| chr3_-_92588982 | 0.84 |
ENSMUST00000098888.5
|
Smcp
|
sperm mitochondria-associated cysteine-rich protein |
| chr10_-_17947997 | 0.84 |
ENSMUST00000037879.6
|
Heca
|
headcase homolog (Drosophila) |
| chr16_-_57606816 | 0.84 |
ENSMUST00000114371.3
|
Cmss1
|
cms small ribosomal subunit 1 |
| chr11_-_6606053 | 0.84 |
ENSMUST00000045713.3
|
Nacad
|
NAC alpha domain containing |
| chr8_+_60993189 | 0.82 |
ENSMUST00000034065.7
ENSMUST00000120689.1 |
Nek1
|
NIMA (never in mitosis gene a)-related expressed kinase 1 |
| chr3_+_89459118 | 0.82 |
ENSMUST00000029564.5
|
Pmvk
|
phosphomevalonate kinase |
| chr9_+_20888175 | 0.82 |
ENSMUST00000004203.5
|
Ppan
|
peter pan homolog (Drosophila) |
| chr6_-_39725193 | 0.80 |
ENSMUST00000101497.3
|
Braf
|
Braf transforming gene |
| chr13_-_49652714 | 0.79 |
ENSMUST00000021818.7
|
Cenpp
|
centromere protein P |
| chr9_+_57827284 | 0.78 |
ENSMUST00000163186.1
|
Gm17231
|
predicted gene 17231 |
| chr7_+_90442729 | 0.78 |
ENSMUST00000061767.4
ENSMUST00000107206.1 |
Crebzf
|
CREB/ATF bZIP transcription factor |
| chr9_+_22411515 | 0.78 |
ENSMUST00000058868.7
|
9530077C05Rik
|
RIKEN cDNA 9530077C05 gene |
| chr4_-_130359915 | 0.78 |
ENSMUST00000134159.2
|
Zcchc17
|
zinc finger, CCHC domain containing 17 |
| chr11_-_69323768 | 0.78 |
ENSMUST00000092973.5
|
Cntrob
|
centrobin, centrosomal BRCA2 interacting protein |
| chr8_+_66860215 | 0.77 |
ENSMUST00000118009.1
|
Naf1
|
nuclear assembly factor 1 homolog (S. cerevisiae) |
| chr2_-_120609283 | 0.77 |
ENSMUST00000102496.1
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr2_-_157007039 | 0.76 |
ENSMUST00000103129.2
ENSMUST00000103130.1 |
Dsn1
|
DSN1, MIND kinetochore complex component, homolog (S. cerevisiae) |
| chr6_-_39725448 | 0.76 |
ENSMUST00000002487.8
|
Braf
|
Braf transforming gene |
| chr7_+_130865835 | 0.75 |
ENSMUST00000075181.4
ENSMUST00000048180.5 |
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chrX_+_11302432 | 0.75 |
ENSMUST00000179428.1
|
Gm14474
|
predicted gene 14474 |
| chr8_-_4325096 | 0.75 |
ENSMUST00000098950.4
|
Elavl1
|
ELAV (embryonic lethal, abnormal vision)-like 1 (Hu antigen R) |
| chr11_-_88718165 | 0.74 |
ENSMUST00000107908.1
|
Msi2
|
musashi RNA-binding protein 2 |
| chr8_-_25597459 | 0.73 |
ENSMUST00000079160.6
|
Letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr2_-_125123618 | 0.73 |
ENSMUST00000142718.1
ENSMUST00000152367.1 ENSMUST00000067780.3 ENSMUST00000147105.1 |
Myef2
|
myelin basic protein expression factor 2, repressor |
| chr15_-_50889691 | 0.71 |
ENSMUST00000165201.2
ENSMUST00000184458.1 |
Trps1
|
trichorhinophalangeal syndrome I (human) |
| chr3_-_158036630 | 0.70 |
ENSMUST00000121326.1
|
Srsf11
|
serine/arginine-rich splicing factor 11 |
| chr7_-_4778141 | 0.70 |
ENSMUST00000094892.5
|
Il11
|
interleukin 11 |
| chr13_-_23761223 | 0.68 |
ENSMUST00000102964.2
|
Hist1h4a
|
histone cluster 1, H4a |
| chr18_-_80440850 | 0.68 |
ENSMUST00000161003.1
|
Ctdp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr6_-_30304513 | 0.67 |
ENSMUST00000094543.2
ENSMUST00000102993.3 |
Ube2h
|
ubiquitin-conjugating enzyme E2H |
| chr1_-_126738167 | 0.67 |
ENSMUST00000160693.1
|
Nckap5
|
NCK-associated protein 5 |
| chr8_-_110997764 | 0.66 |
ENSMUST00000040416.7
|
Ddx19a
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 19a |
| chr11_+_102393403 | 0.65 |
ENSMUST00000107105.2
ENSMUST00000107102.1 ENSMUST00000107103.1 ENSMUST00000006750.7 |
Rundc3a
|
RUN domain containing 3A |
| chr11_-_70237852 | 0.65 |
ENSMUST00000108575.2
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr10_+_79930419 | 0.64 |
ENSMUST00000131118.1
|
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr13_+_3837757 | 0.64 |
ENSMUST00000042219.4
|
Calm4
|
calmodulin 4 |
| chr1_+_71653339 | 0.64 |
ENSMUST00000125934.1
|
Apol7d
|
apolipoprotein L 7d |
| chr3_+_103171655 | 0.64 |
ENSMUST00000005830.8
|
Bcas2
|
breast carcinoma amplified sequence 2 |
| chr3_-_65958236 | 0.63 |
ENSMUST00000029416.7
|
Ccnl1
|
cyclin L1 |
| chr9_-_96478596 | 0.63 |
ENSMUST00000071301.4
|
Rnf7
|
ring finger protein 7 |
| chr1_-_165236892 | 0.62 |
ENSMUST00000043235.5
|
Tiprl
|
TIP41, TOR signalling pathway regulator-like (S. cerevisiae) |
| chr7_+_45718121 | 0.62 |
ENSMUST00000135500.2
|
Rpl18
|
ribosomal protein L18 |
| chr7_+_30712209 | 0.62 |
ENSMUST00000005692.6
ENSMUST00000170371.1 |
Atp4a
|
ATPase, H+/K+ exchanging, gastric, alpha polypeptide |
| chr17_+_70929006 | 0.61 |
ENSMUST00000092011.6
|
Gm16519
|
predicted gene, 16519 |
| chr5_+_146833106 | 0.60 |
ENSMUST00000075453.2
ENSMUST00000099272.2 |
Rpl21
|
ribosomal protein L21 |
| chr5_+_150522599 | 0.60 |
ENSMUST00000044620.7
|
Brca2
|
breast cancer 2 |
| chr5_+_31697960 | 0.60 |
ENSMUST00000114515.2
|
Bre
|
brain and reproductive organ-expressed protein |
| chr2_+_162931520 | 0.59 |
ENSMUST00000130411.1
|
Srsf6
|
serine/arginine-rich splicing factor 6 |
| chr9_-_96478660 | 0.59 |
ENSMUST00000057500.4
|
Rnf7
|
ring finger protein 7 |
| chr19_+_36926071 | 0.59 |
ENSMUST00000099494.3
|
Btaf1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, (Mot1 homolog, S. cerevisiae) |
| chr2_-_157007015 | 0.59 |
ENSMUST00000146413.1
|
Dsn1
|
DSN1, MIND kinetochore complex component, homolog (S. cerevisiae) |
| chr7_-_44748306 | 0.58 |
ENSMUST00000118162.1
ENSMUST00000140599.2 ENSMUST00000120798.1 |
Zfp473
|
zinc finger protein 473 |
| chr17_+_32036098 | 0.58 |
ENSMUST00000081339.6
|
Rrp1b
|
ribosomal RNA processing 1 homolog B (S. cerevisiae) |
| chr7_+_45718058 | 0.58 |
ENSMUST00000072503.6
|
Rpl18
|
ribosomal protein L18 |
| chr11_+_68901538 | 0.58 |
ENSMUST00000073471.6
ENSMUST00000101014.2 ENSMUST00000128952.1 ENSMUST00000167436.1 |
Rpl26
|
ribosomal protein L26 |
| chr1_+_75236439 | 0.58 |
ENSMUST00000082158.6
ENSMUST00000055223.7 |
Dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr4_+_6365650 | 0.57 |
ENSMUST00000029912.4
ENSMUST00000103008.5 ENSMUST00000175769.1 ENSMUST00000108374.1 ENSMUST00000140830.1 |
Sdcbp
|
syndecan binding protein |
| chr14_+_50807915 | 0.56 |
ENSMUST00000036126.5
|
Parp2
|
poly (ADP-ribose) polymerase family, member 2 |
| chr7_+_30422389 | 0.56 |
ENSMUST00000108175.1
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr6_+_41684414 | 0.56 |
ENSMUST00000031900.5
|
1700034O15Rik
|
RIKEN cDNA 1700034O15 gene |
| chr9_-_97111117 | 0.55 |
ENSMUST00000085206.4
|
Slc25a36
|
solute carrier family 25, member 36 |
| chr12_+_79130777 | 0.55 |
ENSMUST00000021550.6
|
Arg2
|
arginase type II |
| chr3_+_67374116 | 0.55 |
ENSMUST00000061322.8
|
Mlf1
|
myeloid leukemia factor 1 |
| chr6_-_149188648 | 0.54 |
ENSMUST00000095319.3
ENSMUST00000141346.1 ENSMUST00000111535.1 |
Amn1
|
antagonist of mitotic exit network 1 |
| chr10_-_128547722 | 0.54 |
ENSMUST00000040572.3
|
Zc3h10
|
zinc finger CCCH type containing 10 |
| chr12_-_110682606 | 0.54 |
ENSMUST00000070659.5
|
1700001K19Rik
|
RIKEN cDNA 1700001K19 gene |
| chr16_+_21794320 | 0.54 |
ENSMUST00000181780.1
ENSMUST00000181960.1 |
1300002E11Rik
|
RIKEN cDNA 1300002E11 gene |
| chr8_+_93810832 | 0.54 |
ENSMUST00000034198.8
ENSMUST00000125716.1 |
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chrX_-_7188713 | 0.53 |
ENSMUST00000004428.7
|
Clcn5
|
chloride channel 5 |
| chr2_-_120609319 | 0.53 |
ENSMUST00000102497.3
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr5_+_142960343 | 0.53 |
ENSMUST00000031565.8
|
Fscn1
|
fascin homolog 1, actin bundling protein (Strongylocentrotus purpuratus) |
| chr14_-_101200069 | 0.53 |
ENSMUST00000057718.4
|
1700110M21Rik
|
RIKEN cDNA 1700110M21 gene |
| chr2_+_24385313 | 0.53 |
ENSMUST00000056641.8
|
Psd4
|
pleckstrin and Sec7 domain containing 4 |
| chr5_-_121191365 | 0.53 |
ENSMUST00000100770.2
ENSMUST00000054547.7 |
Ptpn11
|
protein tyrosine phosphatase, non-receptor type 11 |
| chr17_+_8849974 | 0.52 |
ENSMUST00000115720.1
|
Pde10a
|
phosphodiesterase 10A |
| chr11_-_69900886 | 0.52 |
ENSMUST00000108621.2
ENSMUST00000100969.2 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr7_+_130865756 | 0.52 |
ENSMUST00000120441.1
|
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr3_-_98859753 | 0.52 |
ENSMUST00000029465.6
|
Hsd3b1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| chr7_-_44748617 | 0.52 |
ENSMUST00000060270.6
|
Zfp473
|
zinc finger protein 473 |
| chr5_+_140735526 | 0.51 |
ENSMUST00000120630.2
|
Amz1
|
archaelysin family metallopeptidase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf1_Creb5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.9 | 5.3 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.7 | 3.0 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.7 | 4.9 | GO:0046601 | positive regulation of centriole replication(GO:0046601) de novo centriole assembly(GO:0098535) |
| 0.7 | 2.0 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.7 | 4.0 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.6 | 5.1 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.5 | 1.6 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.5 | 1.9 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 0.4 | 1.6 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.4 | 1.2 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.4 | 1.1 | GO:2000118 | positive regulation of keratinocyte apoptotic process(GO:1902174) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.3 | 1.3 | GO:0006272 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.3 | 1.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.3 | 0.9 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.3 | 0.9 | GO:1904959 | regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.3 | 1.5 | GO:0010920 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) protein secretion by platelet(GO:0070560) |
| 0.3 | 1.4 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.3 | 1.6 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.3 | 0.8 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.3 | 0.8 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.2 | 2.5 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.2 | 2.0 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.2 | 0.8 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.2 | 8.1 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.2 | 3.1 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.2 | 0.6 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.2 | 0.6 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.2 | 3.0 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.2 | 0.6 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.2 | 1.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.2 | 0.5 | GO:2000850 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.2 | 1.7 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.2 | 0.5 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) |
| 0.2 | 0.5 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.2 | 1.5 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 | 3.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 3.2 | GO:0051383 | kinetochore organization(GO:0051383) |
| 0.2 | 0.8 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.2 | 0.9 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.2 | 0.8 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 | 1.6 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 0.4 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.1 | 0.4 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 | 1.7 | GO:0030432 | peristalsis(GO:0030432) |
| 0.1 | 0.4 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 1.0 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.5 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.1 | 0.9 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 | 0.8 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 3.4 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 1.2 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 0.6 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 | 0.3 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.1 | 0.3 | GO:0009196 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.1 | 0.5 | GO:1900108 | sequestering of BMP in extracellular matrix(GO:0035582) negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.6 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.5 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 0.1 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.1 | 0.5 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 | 1.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.4 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.1 | 0.4 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 1.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 0.5 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 1.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 | 0.3 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.1 | 0.6 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.1 | 1.0 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.1 | 0.3 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.6 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 | 0.8 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.9 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 | 0.7 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.3 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 0.3 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 | 3.7 | GO:0070918 | dsRNA fragmentation(GO:0031050) production of miRNAs involved in gene silencing by miRNA(GO:0035196) production of small RNA involved in gene silencing by RNA(GO:0070918) |
| 0.1 | 0.3 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 0.2 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 1.7 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 1.0 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.1 | 0.3 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 2.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.1 | 1.1 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 0.2 | GO:0051309 | female meiosis chromosome separation(GO:0051309) |
| 0.1 | 1.3 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.2 | GO:1904446 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) positive regulation of establishment of Sertoli cell barrier(GO:1904446) negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.4 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 1.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.1 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) |
| 0.1 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.2 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.9 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.8 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 0.6 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.4 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 1.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.4 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 1.3 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 0.6 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 0.1 | 1.9 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 0.4 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.3 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.2 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.2 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) |
| 0.0 | 0.5 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.4 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 2.4 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.3 | GO:0098963 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.0 | 0.4 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 2.0 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.7 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 5.3 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.2 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.2 | GO:0019661 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.3 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.5 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.3 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 1.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.2 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.6 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.0 | 0.6 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.7 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 1.0 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.1 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.4 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.4 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 1.0 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.9 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 2.1 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 1.3 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.2 | GO:0060770 | regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0045423 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.0 | 0.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.7 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.5 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 5.0 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.5 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.0 | 0.2 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.9 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.2 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.5 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 1.4 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.7 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.5 | 6.8 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.4 | 1.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.4 | 1.2 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.4 | 2.9 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.4 | 1.4 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.3 | 1.3 | GO:0000939 | nuclear MIS12/MIND complex(GO:0000818) condensed chromosome inner kinetochore(GO:0000939) |
| 0.3 | 6.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 2.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.2 | 3.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 0.9 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.2 | 1.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.5 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 1.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.5 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.7 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 5.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.4 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 1.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 2.5 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 2.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 1.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.5 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 8.7 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.1 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.5 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 2.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.4 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 3.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0098842 | cytoplasmic side of early endosome membrane(GO:0098559) postsynaptic early endosome(GO:0098842) |
| 0.0 | 1.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.0 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.9 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.2 | GO:0030681 | ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 1.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.6 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.2 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 1.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.5 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 1.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 3.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 4.1 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 3.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 1.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.9 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 2.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 1.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 1.7 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.9 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.8 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.8 | 4.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.8 | 5.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.6 | 6.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.5 | 4.7 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.5 | 3.0 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.4 | 1.3 | GO:0032139 | DNA polymerase processivity factor activity(GO:0030337) dinucleotide insertion or deletion binding(GO:0032139) |
| 0.4 | 1.3 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.4 | 5.1 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.4 | 1.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.3 | 1.7 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.3 | 0.9 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.3 | 1.6 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.2 | 1.0 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.2 | 1.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 3.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 3.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 0.6 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.2 | 0.9 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 2.0 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 1.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.2 | 0.5 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.6 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.4 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.1 | 0.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.6 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 1.0 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 0.3 | GO:0050145 | uridylate kinase activity(GO:0009041) nucleoside phosphate kinase activity(GO:0050145) |
| 0.1 | 0.5 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.1 | 0.9 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.4 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 0.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 2.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 1.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.3 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.3 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.1 | 0.3 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 2.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 1.7 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 0.4 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.1 | 0.5 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 1.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 2.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.5 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.5 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 1.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.2 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 0.2 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) |
| 0.1 | 0.2 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 0.5 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 0.6 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.2 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 1.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.2 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 0.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 5.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 0.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 1.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 1.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.7 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.6 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 1.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 1.2 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 1.8 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 1.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.3 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 1.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 3.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 4.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 1.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 5.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 1.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 1.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.5 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.3 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 3.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 1.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.4 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 1.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 1.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 4.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 1.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 0.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.6 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 3.9 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 4.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.8 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.0 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 2.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 2.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.2 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 3.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.7 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.2 | 2.0 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 4.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 6.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 2.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 6.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 5.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 6.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 3.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 2.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 2.2 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 0.5 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 1.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.8 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 4.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 0.4 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 0.6 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 2.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.3 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 3.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.1 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.0 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 2.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.5 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 1.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.8 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 3.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.8 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.3 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |