Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
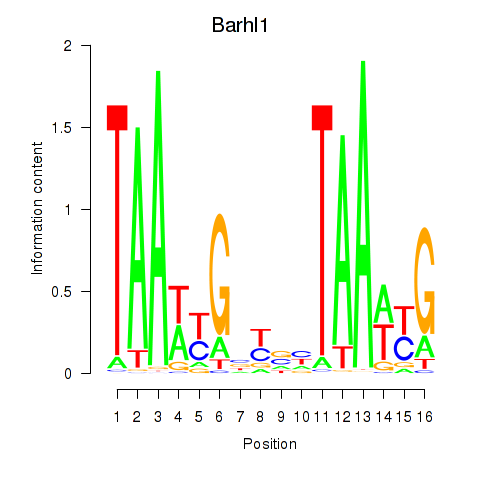
Results for Barhl1
Z-value: 0.46
Transcription factors associated with Barhl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Barhl1
|
ENSMUSG00000026805.8 | BarH like homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Barhl1 | mm10_v2_chr2_-_28916412_28916440 | -0.20 | 2.5e-01 | Click! |
Activity profile of Barhl1 motif
Sorted Z-values of Barhl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_90490714 | 3.99 |
ENSMUST00000042755.3
|
Afp
|
alpha fetoprotein |
| chr17_+_7025837 | 2.58 |
ENSMUST00000089120.5
|
Gm1604b
|
predicted gene 1604b |
| chr17_-_8101228 | 2.25 |
ENSMUST00000097422.4
|
Gm1604A
|
predicted gene 1604A |
| chr3_-_75270073 | 1.70 |
ENSMUST00000039047.4
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr19_-_34166037 | 1.63 |
ENSMUST00000025686.7
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr8_+_31089471 | 1.27 |
ENSMUST00000036631.7
ENSMUST00000170204.1 |
Dusp26
|
dual specificity phosphatase 26 (putative) |
| chr1_-_75278345 | 1.25 |
ENSMUST00000039534.4
|
Resp18
|
regulated endocrine-specific protein 18 |
| chr6_-_129876659 | 1.23 |
ENSMUST00000014687.4
ENSMUST00000122219.1 |
Klra17
|
killer cell lectin-like receptor, subfamily A, member 17 |
| chr18_-_32559914 | 0.87 |
ENSMUST00000174000.1
ENSMUST00000174459.1 |
Gypc
|
glycophorin C |
| chr6_+_123123423 | 0.86 |
ENSMUST00000032248.7
|
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr7_+_30763750 | 0.80 |
ENSMUST00000165887.1
ENSMUST00000085691.4 ENSMUST00000085688.4 ENSMUST00000054427.6 |
Dmkn
|
dermokine |
| chr18_+_53551594 | 0.76 |
ENSMUST00000115398.1
|
Prdm6
|
PR domain containing 6 |
| chrX_-_164250368 | 0.71 |
ENSMUST00000112263.1
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr6_-_41636389 | 0.71 |
ENSMUST00000031902.5
|
Trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr11_+_116198853 | 0.62 |
ENSMUST00000021130.6
|
Ten1
|
TEN1 telomerase capping complex subunit |
| chr2_-_45110336 | 0.52 |
ENSMUST00000028229.6
ENSMUST00000152232.1 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chrX_+_112604274 | 0.51 |
ENSMUST00000071814.6
|
Zfp711
|
zinc finger protein 711 |
| chr6_+_72097561 | 0.51 |
ENSMUST00000069994.4
ENSMUST00000114112.1 |
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr9_+_36832684 | 0.50 |
ENSMUST00000034630.8
|
Fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr7_-_45896656 | 0.48 |
ENSMUST00000120299.1
|
Syngr4
|
synaptogyrin 4 |
| chrX_+_109196750 | 0.47 |
ENSMUST00000139259.1
ENSMUST00000060013.3 |
Gm6377
|
predicted gene 6377 |
| chr8_-_84840627 | 0.46 |
ENSMUST00000003911.6
ENSMUST00000109761.2 ENSMUST00000128035.1 |
Rad23a
|
RAD23a homolog (S. cerevisiae) |
| chr11_+_89073841 | 0.43 |
ENSMUST00000100619.4
|
Gm525
|
predicted gene 525 |
| chr5_+_149678224 | 0.43 |
ENSMUST00000100404.3
|
B3galtl
|
beta 1,3-galactosyltransferase-like |
| chr4_-_117156144 | 0.42 |
ENSMUST00000102696.4
|
Rps8
|
ribosomal protein S8 |
| chr1_-_63214543 | 0.41 |
ENSMUST00000050536.7
|
Gpr1
|
G protein-coupled receptor 1 |
| chr7_+_97842917 | 0.40 |
ENSMUST00000033040.5
|
Pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr15_+_101473472 | 0.40 |
ENSMUST00000088049.3
|
Krt86
|
keratin 86 |
| chr2_-_104849876 | 0.38 |
ENSMUST00000028593.4
|
Prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr6_+_129591782 | 0.35 |
ENSMUST00000112063.2
ENSMUST00000032268.7 ENSMUST00000119520.1 |
Klrd1
|
killer cell lectin-like receptor, subfamily D, member 1 |
| chr2_-_73453918 | 0.33 |
ENSMUST00000102679.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr2_+_122426432 | 0.32 |
ENSMUST00000110525.1
|
Slc28a2
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 2 |
| chr4_+_112232245 | 0.30 |
ENSMUST00000038455.5
ENSMUST00000170945.1 |
Skint3
|
selection and upkeep of intraepithelial T cells 3 |
| chr14_+_25980039 | 0.29 |
ENSMUST00000173155.1
|
Duxbl1
|
double homeobox B-like 1 |
| chr5_-_104077608 | 0.23 |
ENSMUST00000164471.1
ENSMUST00000178967.1 |
Gm17660
|
predicted gene, 17660 |
| chr7_-_45896677 | 0.23 |
ENSMUST00000039049.7
|
Syngr4
|
synaptogyrin 4 |
| chrX_-_75578188 | 0.22 |
ENSMUST00000033545.5
|
Rab39b
|
RAB39B, member RAS oncogene family |
| chr3_-_19265007 | 0.19 |
ENSMUST00000091314.4
|
Pde7a
|
phosphodiesterase 7A |
| chr3_-_130709419 | 0.19 |
ENSMUST00000043937.7
|
Ostc
|
oligosaccharyltransferase complex subunit |
| chr10_+_128747850 | 0.18 |
ENSMUST00000163377.2
|
Wibg
|
within bgcn homolog (Drosophila) |
| chr3_-_19264959 | 0.16 |
ENSMUST00000121951.1
|
Pde7a
|
phosphodiesterase 7A |
| chr5_+_138187485 | 0.16 |
ENSMUST00000110934.2
|
Cnpy4
|
canopy 4 homolog (zebrafish) |
| chr14_+_63860290 | 0.15 |
ENSMUST00000022528.4
|
Pinx1
|
PIN2/TERF1 interacting, telomerase inhibitor 1 |
| chr13_+_19342154 | 0.13 |
ENSMUST00000103566.3
|
Tcrg-C4
|
T cell receptor gamma, constant 4 |
| chr16_+_92292380 | 0.12 |
ENSMUST00000047383.3
|
Kcne2
|
potassium voltage-gated channel, Isk-related subfamily, gene 2 |
| chr18_+_12972225 | 0.11 |
ENSMUST00000025290.5
|
Impact
|
imprinted and ancient |
| chr5_+_34915915 | 0.10 |
ENSMUST00000050535.1
|
Msantd1
|
Myb/SANT-like DNA-binding domain containing 1 |
| chr3_-_88410295 | 0.09 |
ENSMUST00000056370.7
|
Pmf1
|
polyamine-modulated factor 1 |
| chr11_+_78826575 | 0.09 |
ENSMUST00000147875.2
ENSMUST00000141321.1 |
Lyrm9
|
LYR motif containing 9 |
| chr10_+_102159000 | 0.08 |
ENSMUST00000020039.6
|
Mgat4c
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr15_+_81744848 | 0.08 |
ENSMUST00000109554.1
|
Zc3h7b
|
zinc finger CCCH type containing 7B |
| chr2_-_177921815 | 0.08 |
ENSMUST00000108986.2
|
C330013J21Rik
|
RIKEN cDNA C330013J21 gene |
| chr12_-_54795698 | 0.08 |
ENSMUST00000005798.8
|
Snx6
|
sorting nexin 6 |
| chr16_-_29544852 | 0.07 |
ENSMUST00000039090.8
|
Atp13a4
|
ATPase type 13A4 |
| chr18_-_42262053 | 0.07 |
ENSMUST00000097590.3
|
Lars
|
leucyl-tRNA synthetase |
| chr17_+_55892184 | 0.07 |
ENSMUST00000054780.8
|
Zfp959
|
zinc finger protein 959 |
| chr2_+_27165233 | 0.06 |
ENSMUST00000000910.6
|
Dbh
|
dopamine beta hydroxylase |
| chr10_-_127030813 | 0.05 |
ENSMUST00000040560.4
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr10_-_127030789 | 0.04 |
ENSMUST00000120547.1
ENSMUST00000152054.1 |
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr10_+_102158858 | 0.03 |
ENSMUST00000138522.1
ENSMUST00000163753.1 ENSMUST00000138016.1 |
Mgat4c
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr1_+_115684727 | 0.02 |
ENSMUST00000043725.7
|
Cntnap5a
|
contactin associated protein-like 5A |
| chr2_-_80128834 | 0.02 |
ENSMUST00000102654.4
ENSMUST00000102655.3 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr17_-_25115905 | 0.02 |
ENSMUST00000024987.5
ENSMUST00000115181.2 |
Telo2
|
TEL2, telomere maintenance 2, homolog (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Barhl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.2 | 4.0 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 0.8 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.5 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.4 | GO:0021764 | amygdala development(GO:0021764) negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 0.3 | GO:1901642 | purine nucleoside transmembrane transport(GO:0015860) nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 0.7 | GO:1990035 | parathyroid hormone secretion(GO:0035898) calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.8 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 0.2 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.7 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.6 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.5 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.1 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.5 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.5 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 0.6 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.4 | 1.3 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.3 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 0.3 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.1 | 1.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.5 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.5 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |