Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
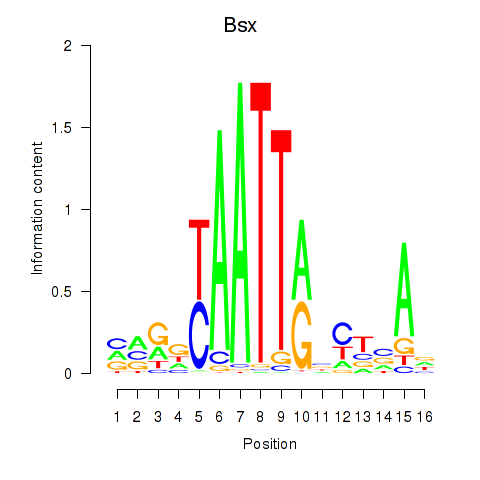
Results for Bsx
Z-value: 0.58
Transcription factors associated with Bsx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bsx
|
ENSMUSG00000054360.3 | brain specific homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bsx | mm10_v2_chr9_+_40873981_40874127 | 0.23 | 1.8e-01 | Click! |
Activity profile of Bsx motif
Sorted Z-values of Bsx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_119459266 | 3.74 |
ENSMUST00000033255.5
|
Gp2
|
glycoprotein 2 (zymogen granule membrane) |
| chr11_+_58640394 | 2.44 |
ENSMUST00000075084.4
|
Trim58
|
tripartite motif-containing 58 |
| chr5_-_137072254 | 2.17 |
ENSMUST00000077523.3
ENSMUST00000041388.4 |
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1 |
| chr1_+_40439767 | 1.91 |
ENSMUST00000173514.1
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr2_-_113829069 | 1.84 |
ENSMUST00000024005.7
|
Scg5
|
secretogranin V |
| chr3_-_116253467 | 1.47 |
ENSMUST00000090473.5
|
Gpr88
|
G-protein coupled receptor 88 |
| chr17_-_47833169 | 1.08 |
ENSMUST00000131971.1
ENSMUST00000129360.1 ENSMUST00000113280.1 ENSMUST00000132125.1 |
Mdfi
|
MyoD family inhibitor |
| chr2_-_32387760 | 1.06 |
ENSMUST00000050785.8
|
Lcn2
|
lipocalin 2 |
| chr1_+_40439627 | 1.02 |
ENSMUST00000097772.3
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr17_-_47833256 | 1.02 |
ENSMUST00000152455.1
ENSMUST00000035375.7 |
Mdfi
|
MyoD family inhibitor |
| chr19_-_56389877 | 0.95 |
ENSMUST00000166203.1
ENSMUST00000167239.1 ENSMUST00000040711.8 ENSMUST00000095947.4 ENSMUST00000073536.6 |
Nrap
|
nebulin-related anchoring protein |
| chr4_-_115133977 | 0.87 |
ENSMUST00000051400.7
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr6_+_29694204 | 0.87 |
ENSMUST00000046750.7
ENSMUST00000115250.3 |
Tspan33
|
tetraspanin 33 |
| chr13_-_23745511 | 0.84 |
ENSMUST00000091752.2
|
Hist1h3c
|
histone cluster 1, H3c |
| chr3_-_15575065 | 0.83 |
ENSMUST00000091319.4
|
Sirpb1b
|
signal-regulatory protein beta 1B |
| chr16_+_52031549 | 0.78 |
ENSMUST00000114471.1
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr11_-_98053415 | 0.69 |
ENSMUST00000017544.2
|
Stac2
|
SH3 and cysteine rich domain 2 |
| chr2_+_69219971 | 0.68 |
ENSMUST00000005364.5
ENSMUST00000112317.2 |
G6pc2
|
glucose-6-phosphatase, catalytic, 2 |
| chr12_+_35992900 | 0.64 |
ENSMUST00000020898.5
|
Agr2
|
anterior gradient 2 |
| chr17_+_34039437 | 0.63 |
ENSMUST00000131134.1
ENSMUST00000087497.4 ENSMUST00000114255.1 ENSMUST00000114252.1 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr8_+_57511833 | 0.62 |
ENSMUST00000067925.6
|
Hmgb2
|
high mobility group box 2 |
| chr17_-_31277327 | 0.60 |
ENSMUST00000024832.7
|
Rsph1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr19_+_4594312 | 0.55 |
ENSMUST00000113825.2
|
Pcx
|
pyruvate carboxylase |
| chr9_-_95750335 | 0.54 |
ENSMUST00000053785.3
|
Trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr6_-_50456085 | 0.52 |
ENSMUST00000146341.1
ENSMUST00000071728.4 |
Osbpl3
|
oxysterol binding protein-like 3 |
| chr4_-_41464816 | 0.52 |
ENSMUST00000108055.2
ENSMUST00000154535.1 ENSMUST00000030148.5 |
Kif24
|
kinesin family member 24 |
| chr15_+_92597104 | 0.46 |
ENSMUST00000035399.8
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr7_+_35802593 | 0.45 |
ENSMUST00000052454.2
|
E130304I02Rik
|
RIKEN cDNA E130304I02 gene |
| chr2_-_110362985 | 0.45 |
ENSMUST00000099626.3
|
Fibin
|
fin bud initiation factor homolog (zebrafish) |
| chr1_+_130731963 | 0.44 |
ENSMUST00000039323.6
|
AA986860
|
expressed sequence AA986860 |
| chr7_+_3645267 | 0.43 |
ENSMUST00000038913.9
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr2_+_92375306 | 0.43 |
ENSMUST00000028650.8
|
Pex16
|
peroxisomal biogenesis factor 16 |
| chr5_+_136987019 | 0.42 |
ENSMUST00000004968.4
|
Plod3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr3_-_15848419 | 0.42 |
ENSMUST00000108354.1
ENSMUST00000108349.1 ENSMUST00000108352.2 ENSMUST00000108350.1 ENSMUST00000050623.4 |
Sirpb1c
|
signal-regulatory protein beta 1C |
| chr2_-_120154600 | 0.41 |
ENSMUST00000028755.7
|
Ehd4
|
EH-domain containing 4 |
| chr2_+_110721587 | 0.40 |
ENSMUST00000111017.2
|
Muc15
|
mucin 15 |
| chr6_-_124779686 | 0.40 |
ENSMUST00000147669.1
ENSMUST00000128697.1 ENSMUST00000032218.3 ENSMUST00000112475.2 |
Lrrc23
|
leucine rich repeat containing 23 |
| chr17_+_24886643 | 0.40 |
ENSMUST00000117890.1
ENSMUST00000168265.1 ENSMUST00000120943.1 ENSMUST00000068508.6 ENSMUST00000119829.1 |
Spsb3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr8_-_77517898 | 0.39 |
ENSMUST00000076316.4
|
Arhgap10
|
Rho GTPase activating protein 10 |
| chr5_-_103977404 | 0.37 |
ENSMUST00000112803.2
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr5_-_103977326 | 0.36 |
ENSMUST00000120320.1
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr2_+_110721340 | 0.33 |
ENSMUST00000111016.2
|
Muc15
|
mucin 15 |
| chr19_-_55241236 | 0.33 |
ENSMUST00000069183.6
|
Gucy2g
|
guanylate cyclase 2g |
| chr18_+_77332394 | 0.33 |
ENSMUST00000148341.1
|
Loxhd1
|
lipoxygenase homology domains 1 |
| chr5_-_103977360 | 0.32 |
ENSMUST00000048118.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr6_+_72097561 | 0.30 |
ENSMUST00000069994.4
ENSMUST00000114112.1 |
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr14_-_108914237 | 0.28 |
ENSMUST00000100322.2
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr2_+_85979312 | 0.28 |
ENSMUST00000170610.1
|
Olfr1030
|
olfactory receptor 1030 |
| chr17_-_24886304 | 0.27 |
ENSMUST00000044252.5
|
Nubp2
|
nucleotide binding protein 2 |
| chr2_+_116067213 | 0.26 |
ENSMUST00000152412.1
|
G630016G05Rik
|
RIKEN cDNA G630016G05 gene |
| chrX_+_56786527 | 0.24 |
ENSMUST00000144600.1
|
Fhl1
|
four and a half LIM domains 1 |
| chr5_+_33335540 | 0.23 |
ENSMUST00000114449.3
|
Maea
|
macrophage erythroblast attacher |
| chr5_+_103425181 | 0.22 |
ENSMUST00000048957.9
|
Ptpn13
|
protein tyrosine phosphatase, non-receptor type 13 |
| chr15_-_8710409 | 0.22 |
ENSMUST00000157065.1
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr11_+_90883320 | 0.21 |
ENSMUST00000142163.1
ENSMUST00000132218.1 |
4930405D11Rik
|
RIKEN cDNA 4930405D11 gene |
| chr2_+_3114220 | 0.21 |
ENSMUST00000072955.5
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr5_+_134932351 | 0.20 |
ENSMUST00000047196.7
ENSMUST00000111221.2 ENSMUST00000111219.1 ENSMUST00000068617.5 ENSMUST00000111218.1 ENSMUST00000136246.1 |
Wbscr27
|
Williams Beuren syndrome chromosome region 27 (human) |
| chr12_+_72441852 | 0.19 |
ENSMUST00000162159.1
|
Lrrc9
|
leucine rich repeat containing 9 |
| chr5_-_136986829 | 0.19 |
ENSMUST00000034953.7
ENSMUST00000085941.5 |
Znhit1
|
zinc finger, HIT domain containing 1 |
| chr12_-_112802646 | 0.17 |
ENSMUST00000124526.1
|
Ahnak2
|
AHNAK nucleoprotein 2 |
| chr5_-_122989260 | 0.17 |
ENSMUST00000118027.1
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr5_+_144100387 | 0.16 |
ENSMUST00000041804.7
|
Lmtk2
|
lemur tyrosine kinase 2 |
| chr10_-_53699343 | 0.15 |
ENSMUST00000163761.1
|
Fam184a
|
family with sequence similarity 184, member A |
| chr2_+_36230426 | 0.15 |
ENSMUST00000062069.5
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr7_+_101896340 | 0.13 |
ENSMUST00000035395.7
ENSMUST00000106973.1 ENSMUST00000144207.1 |
Anapc15
|
anaphase prompoting complex C subunit 15 |
| chr5_+_138187485 | 0.13 |
ENSMUST00000110934.2
|
Cnpy4
|
canopy 4 homolog (zebrafish) |
| chr2_-_67194695 | 0.13 |
ENSMUST00000147939.1
|
Gm13598
|
predicted gene 13598 |
| chr11_-_70646972 | 0.12 |
ENSMUST00000014750.8
|
Slc25a11
|
solute carrier family 25 (mitochondrial carrier oxoglutarate carrier), member 11 |
| chr7_+_25077205 | 0.12 |
ENSMUST00000179556.1
ENSMUST00000053410.9 |
Zfp574
|
zinc finger protein 574 |
| chr13_+_21716385 | 0.12 |
ENSMUST00000070124.3
|
Hist1h2ai
|
histone cluster 1, H2ai |
| chr11_+_116843278 | 0.11 |
ENSMUST00000106370.3
|
Mettl23
|
methyltransferase like 23 |
| chrX_+_13632769 | 0.11 |
ENSMUST00000096492.3
|
Gpr34
|
G protein-coupled receptor 34 |
| chr6_+_63255971 | 0.10 |
ENSMUST00000159561.1
ENSMUST00000095852.3 |
Grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr3_+_55782500 | 0.09 |
ENSMUST00000075422.4
|
Mab21l1
|
mab-21-like 1 (C. elegans) |
| chr12_-_55014329 | 0.09 |
ENSMUST00000172875.1
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr12_+_72441933 | 0.09 |
ENSMUST00000161284.1
|
Lrrc9
|
leucine rich repeat containing 9 |
| chr1_+_171214013 | 0.08 |
ENSMUST00000111328.1
|
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr6_-_34317442 | 0.08 |
ENSMUST00000154655.1
ENSMUST00000102980.4 |
Akr1b3
|
aldo-keto reductase family 1, member B3 (aldose reductase) |
| chr7_+_101896817 | 0.07 |
ENSMUST00000143835.1
|
Anapc15
|
anaphase prompoting complex C subunit 15 |
| chr16_+_34690548 | 0.07 |
ENSMUST00000023532.6
|
Ccdc14
|
coiled-coil domain containing 14 |
| chr4_-_110286581 | 0.06 |
ENSMUST00000138972.1
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D) |
| chrY_-_41644045 | 0.06 |
ENSMUST00000180058.1
|
Gm21729
|
predicted gene, 21729 |
| chr13_-_105054895 | 0.05 |
ENSMUST00000063551.5
|
Rgs7bp
|
regulator of G-protein signalling 7 binding protein |
| chr16_-_63864114 | 0.05 |
ENSMUST00000064405.6
|
Epha3
|
Eph receptor A3 |
| chr5_-_138187177 | 0.05 |
ENSMUST00000110937.1
ENSMUST00000139276.1 ENSMUST00000048698.7 ENSMUST00000123415.1 |
Taf6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr3_+_54361103 | 0.04 |
ENSMUST00000107985.3
ENSMUST00000117373.1 ENSMUST00000073012.6 ENSMUST00000081564.6 |
Postn
|
periostin, osteoblast specific factor |
| chr7_+_103979176 | 0.04 |
ENSMUST00000098185.1
|
Olfr635
|
olfactory receptor 635 |
| chr2_-_116067391 | 0.04 |
ENSMUST00000140185.1
|
2700033N17Rik
|
RIKEN cDNA 2700033N17 gene |
| chr3_-_15426427 | 0.03 |
ENSMUST00000099201.3
|
Sirpb1a
|
signal-regulatory protein beta 1A |
| chr6_+_57002300 | 0.03 |
ENSMUST00000079669.4
|
Vmn1r6
|
vomeronasal 1 receptor 6 |
| chr1_-_163725123 | 0.03 |
ENSMUST00000159679.1
|
Mettl11b
|
methyltransferase like 11B |
| chr4_+_62525369 | 0.02 |
ENSMUST00000062145.1
|
4933430I17Rik
|
RIKEN cDNA 4933430I17 gene |
| chr4_-_91376433 | 0.02 |
ENSMUST00000107109.2
ENSMUST00000107111.2 ENSMUST00000107120.1 |
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B) |
| chrY_+_43953619 | 0.02 |
ENSMUST00000179551.1
|
Gm21241
|
predicted gene, 21241 |
| chr17_+_71781947 | 0.02 |
ENSMUST00000024854.7
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr3_+_59952185 | 0.01 |
ENSMUST00000094227.3
|
Gm9696
|
predicted gene 9696 |
| chrY_+_52781462 | 0.01 |
ENSMUST00000178673.1
|
Gm21258
|
predicted gene, 21258 |
| chr12_+_38780284 | 0.01 |
ENSMUST00000162563.1
ENSMUST00000161164.1 ENSMUST00000160996.1 |
Etv1
|
ets variant gene 1 |
| chrY_+_29351845 | 0.01 |
ENSMUST00000180071.1
|
Gm21853
|
predicted gene, 21853 |
| chr15_+_98092569 | 0.01 |
ENSMUST00000163507.1
|
Pfkm
|
phosphofructokinase, muscle |
| chr15_+_78926720 | 0.00 |
ENSMUST00000089377.5
|
Lgals1
|
lectin, galactose binding, soluble 1 |
| chr19_+_11965817 | 0.00 |
ENSMUST00000025590.9
|
Osbp
|
oxysterol binding protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Bsx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.7 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.7 | 2.2 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.4 | 1.5 | GO:0061743 | motor learning(GO:0061743) |
| 0.3 | 2.9 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.2 | 0.5 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 | 0.6 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.2 | 0.8 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.2 | 1.1 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.1 | 0.4 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.6 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.6 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 2.1 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 0.4 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.1 | 1.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.0 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.5 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.2 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.7 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.1 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0035633 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.2 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.4 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.4 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 1.0 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.9 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 3.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.8 | GO:0000786 | nucleosome(GO:0000786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.2 | 0.7 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.4 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 0.5 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 1.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 3.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.9 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 3.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |