Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
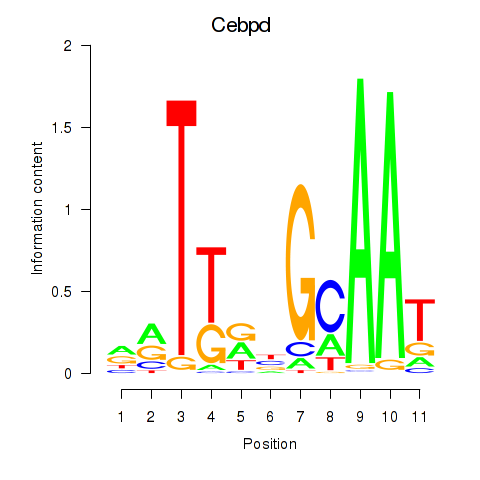
Results for Cebpd
Z-value: 1.27
Transcription factors associated with Cebpd
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cebpd
|
ENSMUSG00000071637.4 | CCAAT/enhancer binding protein (C/EBP), delta |
Activity profile of Cebpd motif
Sorted Z-values of Cebpd motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_46751832 | 20.13 |
ENSMUST00000075982.2
|
Saa2
|
serum amyloid A 2 |
| chr4_+_63344548 | 18.37 |
ENSMUST00000030044.2
|
Orm1
|
orosomucoid 1 |
| chr4_+_63362443 | 17.91 |
ENSMUST00000075341.3
|
Orm2
|
orosomucoid 2 |
| chr9_-_48605147 | 15.45 |
ENSMUST00000034808.5
ENSMUST00000119426.1 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr17_+_36942910 | 12.07 |
ENSMUST00000040498.5
|
Rnf39
|
ring finger protein 39 |
| chr17_+_36943025 | 10.38 |
ENSMUST00000173072.1
|
Rnf39
|
ring finger protein 39 |
| chrX_+_59999436 | 8.89 |
ENSMUST00000033477.4
|
F9
|
coagulation factor IX |
| chr17_-_56121946 | 8.26 |
ENSMUST00000041357.7
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr5_+_114146525 | 6.13 |
ENSMUST00000102582.1
|
Acacb
|
acetyl-Coenzyme A carboxylase beta |
| chr11_-_75422524 | 5.92 |
ENSMUST00000125982.1
ENSMUST00000137103.1 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr10_+_93488766 | 5.78 |
ENSMUST00000129421.1
|
Hal
|
histidine ammonia lyase |
| chr6_-_124542281 | 5.72 |
ENSMUST00000159463.1
ENSMUST00000162844.1 ENSMUST00000160505.1 ENSMUST00000162443.1 |
C1s
|
complement component 1, s subcomponent |
| chr11_-_75422586 | 5.29 |
ENSMUST00000138661.1
ENSMUST00000000769.7 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr6_+_17463749 | 5.06 |
ENSMUST00000115443.1
|
Met
|
met proto-oncogene |
| chr1_-_162898665 | 5.01 |
ENSMUST00000111510.1
ENSMUST00000045902.6 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr18_+_20944607 | 4.53 |
ENSMUST00000050004.1
|
Rnf125
|
ring finger protein 125 |
| chr1_-_140183404 | 3.95 |
ENSMUST00000066859.6
ENSMUST00000111976.2 |
Cfh
|
complement component factor h |
| chr1_-_140183283 | 3.90 |
ENSMUST00000111977.1
|
Cfh
|
complement component factor h |
| chr9_-_103230415 | 3.41 |
ENSMUST00000035158.9
|
Trf
|
transferrin |
| chr7_-_127273919 | 3.39 |
ENSMUST00000082428.3
|
Sephs2
|
selenophosphate synthetase 2 |
| chr5_-_147894804 | 3.17 |
ENSMUST00000118527.1
ENSMUST00000031655.3 ENSMUST00000138244.1 |
Slc46a3
|
solute carrier family 46, member 3 |
| chr6_-_3968357 | 2.98 |
ENSMUST00000031674.8
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr9_-_103230262 | 2.98 |
ENSMUST00000165296.1
ENSMUST00000112645.1 ENSMUST00000166836.1 |
Trf
Gm20425
|
transferrin predicted gene 20425 |
| chr17_-_85090204 | 2.72 |
ENSMUST00000072406.3
ENSMUST00000171795.1 |
Prepl
|
prolyl endopeptidase-like |
| chr5_+_7960445 | 2.62 |
ENSMUST00000115421.1
|
Steap4
|
STEAP family member 4 |
| chrX_+_94234594 | 2.61 |
ENSMUST00000153900.1
|
Klhl15
|
kelch-like 15 |
| chr3_+_19957088 | 2.59 |
ENSMUST00000108328.1
|
Cp
|
ceruloplasmin |
| chr11_-_77894096 | 2.55 |
ENSMUST00000017597.4
|
Pipox
|
pipecolic acid oxidase |
| chr3_+_19957037 | 2.51 |
ENSMUST00000091309.5
ENSMUST00000108329.1 ENSMUST00000003714.6 |
Cp
|
ceruloplasmin |
| chr16_-_23029062 | 2.27 |
ENSMUST00000115349.2
|
Kng2
|
kininogen 2 |
| chr16_-_23029012 | 2.16 |
ENSMUST00000039338.6
|
Kng2
|
kininogen 2 |
| chr16_-_23029080 | 2.12 |
ENSMUST00000100046.2
|
Kng2
|
kininogen 2 |
| chr11_-_106216318 | 2.11 |
ENSMUST00000002043.3
|
Ccdc47
|
coiled-coil domain containing 47 |
| chr3_+_19957240 | 2.06 |
ENSMUST00000108325.2
|
Cp
|
ceruloplasmin |
| chr14_-_45477856 | 2.06 |
ENSMUST00000141424.1
|
Fermt2
|
fermitin family homolog 2 (Drosophila) |
| chr14_-_18893623 | 2.06 |
ENSMUST00000177259.1
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr12_+_37241633 | 1.99 |
ENSMUST00000049874.7
|
Agmo
|
alkylglycerol monooxygenase |
| chr14_-_68533689 | 1.98 |
ENSMUST00000022640.7
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr14_+_75242287 | 1.93 |
ENSMUST00000022576.8
|
Cpb2
|
carboxypeptidase B2 (plasma) |
| chrX_-_162565514 | 1.91 |
ENSMUST00000154424.1
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr7_-_46715676 | 1.83 |
ENSMUST00000006956.7
|
Saa3
|
serum amyloid A 3 |
| chr6_+_29529277 | 1.80 |
ENSMUST00000163511.1
|
Irf5
|
interferon regulatory factor 5 |
| chr4_+_55350043 | 1.75 |
ENSMUST00000030134.8
|
Rad23b
|
RAD23b homolog (S. cerevisiae) |
| chr12_+_37241729 | 1.71 |
ENSMUST00000160768.1
|
Agmo
|
alkylglycerol monooxygenase |
| chr8_+_94037198 | 1.66 |
ENSMUST00000109556.2
ENSMUST00000093301.2 ENSMUST00000060632.7 |
Ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr12_+_24572276 | 1.57 |
ENSMUST00000085553.5
|
Grhl1
|
grainyhead-like 1 (Drosophila) |
| chr14_-_18893376 | 1.57 |
ENSMUST00000151926.1
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr13_+_63015167 | 1.56 |
ENSMUST00000021911.8
|
2010111I01Rik
|
RIKEN cDNA 2010111I01 gene |
| chr4_-_108833608 | 1.53 |
ENSMUST00000102742.4
|
Btf3l4
|
basic transcription factor 3-like 4 |
| chr11_-_78080360 | 1.40 |
ENSMUST00000021183.3
|
Eral1
|
Era (G-protein)-like 1 (E. coli) |
| chr13_+_16011851 | 1.38 |
ENSMUST00000042603.6
|
Inhba
|
inhibin beta-A |
| chr13_+_63014934 | 1.36 |
ENSMUST00000091560.4
|
2010111I01Rik
|
RIKEN cDNA 2010111I01 gene |
| chr5_+_63812447 | 1.29 |
ENSMUST00000081747.3
|
0610040J01Rik
|
RIKEN cDNA 0610040J01 gene |
| chr12_+_86421628 | 1.24 |
ENSMUST00000110203.1
ENSMUST00000110204.2 |
Esrrb
|
estrogen related receptor, beta |
| chr10_-_86011833 | 1.18 |
ENSMUST00000105304.1
ENSMUST00000061699.5 |
Bpifc
|
BPI fold containing family C |
| chr10_+_67535465 | 1.15 |
ENSMUST00000145754.1
|
Egr2
|
early growth response 2 |
| chr2_+_136501978 | 0.98 |
ENSMUST00000061569.2
|
BC034902
|
cDNA sequence BC034902 |
| chr8_-_33929798 | 0.83 |
ENSMUST00000033995.7
ENSMUST00000033994.8 ENSMUST00000053251.5 |
Rbpms
|
RNA binding protein gene with multiple splicing |
| chrX_+_7763943 | 0.83 |
ENSMUST00000144900.1
ENSMUST00000115677.1 ENSMUST00000101695.2 ENSMUST00000115678.2 |
Tfe3
|
transcription factor E3 |
| chr10_+_67535493 | 0.80 |
ENSMUST00000145936.1
|
Egr2
|
early growth response 2 |
| chr7_-_16244152 | 0.76 |
ENSMUST00000171425.1
|
C5ar2
|
complement component 5a receptor 2 |
| chr2_+_3513035 | 0.74 |
ENSMUST00000036350.2
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr1_-_107161114 | 0.72 |
ENSMUST00000086694.4
|
Serpinb3b
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3B |
| chr5_+_115279666 | 0.71 |
ENSMUST00000040421.4
|
Coq5
|
coenzyme Q5 homolog, methyltransferase (yeast) |
| chr5_-_145140238 | 0.69 |
ENSMUST00000031627.8
|
Pdap1
|
PDGFA associated protein 1 |
| chr10_+_33863935 | 0.68 |
ENSMUST00000092597.3
|
Sult3a1
|
sulfotransferase family 3A, member 1 |
| chr17_+_19362135 | 0.67 |
ENSMUST00000176107.1
|
Vmn2r99
|
vomeronasal 2, receptor 99 |
| chr4_-_108833544 | 0.58 |
ENSMUST00000102740.1
ENSMUST00000102741.1 |
Btf3l4
|
basic transcription factor 3-like 4 |
| chr2_-_41789078 | 0.57 |
ENSMUST00000167270.2
|
Lrp1b
|
low density lipoprotein-related protein 1B (deleted in tumors) |
| chr3_-_89913144 | 0.56 |
ENSMUST00000029559.6
|
Il6ra
|
interleukin 6 receptor, alpha |
| chr11_+_6415443 | 0.53 |
ENSMUST00000132846.1
|
Ppia
|
peptidylprolyl isomerase A |
| chr1_+_71653339 | 0.46 |
ENSMUST00000125934.1
|
Apol7d
|
apolipoprotein L 7d |
| chr11_-_48826655 | 0.46 |
ENSMUST00000104959.1
|
Gm12184
|
predicted gene 12184 |
| chr5_+_145140362 | 0.45 |
ENSMUST00000162594.1
ENSMUST00000162308.1 ENSMUST00000159018.1 ENSMUST00000160075.1 |
Bud31
|
BUD31 homolog (yeast) |
| chr3_+_66219909 | 0.43 |
ENSMUST00000029421.5
|
Ptx3
|
pentraxin related gene |
| chr6_-_87809757 | 0.36 |
ENSMUST00000032134.7
|
Rab43
|
RAB43, member RAS oncogene family |
| chr19_-_57239310 | 0.33 |
ENSMUST00000111559.1
|
Ablim1
|
actin-binding LIM protein 1 |
| chr3_+_154762157 | 0.33 |
ENSMUST00000098496.3
|
BC007180
|
cDNA sequence BC007180 |
| chr11_-_87086752 | 0.29 |
ENSMUST00000020801.7
|
Smg8
|
smg-8 homolog, nonsense mediated mRNA decay factor (C. elegans) |
| chr2_-_129371131 | 0.24 |
ENSMUST00000028881.7
|
Il1b
|
interleukin 1 beta |
| chr2_-_57113053 | 0.21 |
ENSMUST00000112627.1
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr1_-_107052331 | 0.20 |
ENSMUST00000112717.1
|
Serpinb3a
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3A |
| chr9_+_99456243 | 0.11 |
ENSMUST00000163199.2
|
1600029I14Rik
|
RIKEN cDNA 1600029I14 gene |
| chr19_+_8819401 | 0.10 |
ENSMUST00000096753.3
|
Hnrnpul2
|
heterogeneous nuclear ribonucleoprotein U-like 2 |
| chr11_-_48826500 | 0.08 |
ENSMUST00000161192.2
|
Gm12184
|
predicted gene 12184 |
| chr2_+_139493913 | 0.07 |
ENSMUST00000110083.1
ENSMUST00000047370.2 |
Sptlc3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr6_+_92869357 | 0.06 |
ENSMUST00000113434.1
|
Gm15737
|
predicted gene 15737 |
| chr9_-_15112100 | 0.05 |
ENSMUST00000159985.1
|
Hephl1
|
hephaestin-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cebpd
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.1 | GO:2001293 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 1.2 | 5.8 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 1.0 | 58.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 1.0 | 11.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 1.0 | 7.8 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.8 | 2.5 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.8 | 3.4 | GO:0016259 | selenium compound metabolic process(GO:0001887) selenocysteine metabolic process(GO:0016259) |
| 0.8 | 5.0 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.8 | 6.4 | GO:0097460 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.7 | 5.1 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.6 | 1.9 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.6 | 1.8 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.5 | 2.0 | GO:0021664 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.5 | 3.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.4 | 2.6 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.3 | 1.2 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.3 | 9.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.3 | 1.4 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.2 | 3.7 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 8.3 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.2 | 2.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.2 | 0.8 | GO:0071623 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 0.2 | 0.6 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.2 | 1.7 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 0.4 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 0.4 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 6.5 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 1.6 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 2.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 2.7 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 | 8.9 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.1 | 0.2 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 1.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.8 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.8 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 4.5 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.7 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 15.2 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.7 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 2.1 | GO:0048041 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 | 0.7 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.7 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.8 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 11.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.6 | 6.4 | GO:0097433 | dense body(GO:0097433) |
| 0.6 | 22.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.5 | 1.4 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.4 | 1.8 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 0.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 19.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 5.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 2.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.8 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 2.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 41.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 2.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.1 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 1.2 | 3.7 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 1.2 | 6.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 1.1 | 7.8 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.9 | 6.4 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.8 | 5.8 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.7 | 7.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.7 | 22.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.6 | 2.5 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.6 | 3.6 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.5 | 5.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 2.6 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 1.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 0.8 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.2 | 0.6 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.2 | 0.8 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.2 | 15.2 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 2.7 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 1.7 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.1 | 8.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 2.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 13.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 4.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 2.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 8.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.2 | GO:0001223 | RNA polymerase II core binding(GO:0000993) transcription coactivator binding(GO:0001223) |
| 0.0 | 0.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 2.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.8 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 2.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 1.6 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 7.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 17.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 7.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.8 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.7 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.6 | 8.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.6 | 7.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 6.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 7.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 15.5 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.2 | 1.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 0.5 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 5.1 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 4.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 5.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 2.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.8 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 5.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 2.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 3.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |