Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
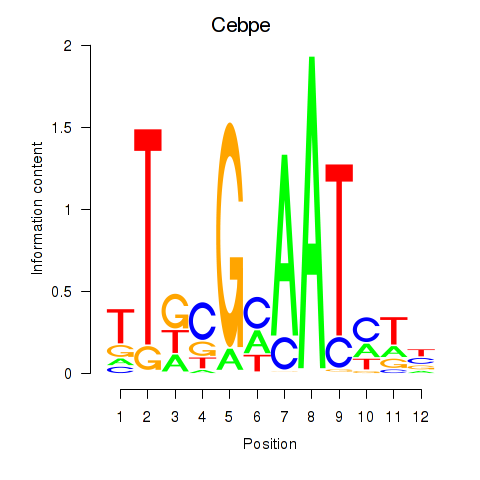
Results for Cebpe
Z-value: 2.96
Transcription factors associated with Cebpe
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cebpe
|
ENSMUSG00000052435.6 | CCAAT/enhancer binding protein (C/EBP), epsilon |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cebpe | mm10_v2_chr14_-_54712139_54712174 | -0.77 | 5.0e-08 | Click! |
Activity profile of Cebpe motif
Sorted Z-values of Cebpe motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_128960965 | 36.79 |
ENSMUST00000026398.3
|
Mettl7b
|
methyltransferase like 7B |
| chr5_-_87337165 | 32.14 |
ENSMUST00000031195.2
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr4_-_60499332 | 31.88 |
ENSMUST00000135953.1
|
Mup1
|
major urinary protein 1 |
| chr1_-_139781236 | 31.43 |
ENSMUST00000027612.8
ENSMUST00000111989.2 ENSMUST00000111986.2 |
Gm4788
|
predicted gene 4788 |
| chr19_-_39463067 | 29.98 |
ENSMUST00000035488.2
|
Cyp2c38
|
cytochrome P450, family 2, subfamily c, polypeptide 38 |
| chr19_+_39510844 | 29.23 |
ENSMUST00000025968.4
|
Cyp2c39
|
cytochrome P450, family 2, subfamily c, polypeptide 39 |
| chr19_+_39287074 | 28.02 |
ENSMUST00000003137.8
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr15_+_9335550 | 24.29 |
ENSMUST00000072403.6
|
Ugt3a2
|
UDP glycosyltransferases 3 family, polypeptide A2 |
| chr8_-_72212837 | 20.39 |
ENSMUST00000098630.3
|
Cib3
|
calcium and integrin binding family member 3 |
| chr6_-_55175019 | 18.94 |
ENSMUST00000003569.5
|
Inmt
|
indolethylamine N-methyltransferase |
| chr7_+_46751832 | 18.74 |
ENSMUST00000075982.2
|
Saa2
|
serum amyloid A 2 |
| chr5_-_87254804 | 18.58 |
ENSMUST00000075858.3
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr5_-_87424201 | 17.42 |
ENSMUST00000072818.5
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr19_-_8131982 | 17.14 |
ENSMUST00000065651.4
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr10_+_128254131 | 16.56 |
ENSMUST00000060782.3
|
Apon
|
apolipoprotein N |
| chr5_-_87140318 | 16.39 |
ENSMUST00000067790.6
ENSMUST00000113327.1 |
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr4_-_60582152 | 16.32 |
ENSMUST00000098047.2
|
Mup10
|
major urinary protein 10 |
| chr9_-_48605147 | 15.80 |
ENSMUST00000034808.5
ENSMUST00000119426.1 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr5_-_87092546 | 15.69 |
ENSMUST00000132667.1
ENSMUST00000145617.1 ENSMUST00000094649.4 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr4_-_60662358 | 15.42 |
ENSMUST00000084544.4
ENSMUST00000098046.3 |
Mup11
|
major urinary protein 11 |
| chr19_-_7802578 | 14.80 |
ENSMUST00000120522.1
ENSMUST00000065634.7 |
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr4_-_60139857 | 14.17 |
ENSMUST00000107490.4
ENSMUST00000074700.2 |
Mup2
|
major urinary protein 2 |
| chr6_-_141856171 | 14.16 |
ENSMUST00000165990.1
ENSMUST00000163678.1 |
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr19_+_39992424 | 13.79 |
ENSMUST00000049178.2
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr4_-_61674094 | 13.46 |
ENSMUST00000098040.3
|
Mup18
|
major urinary protein 18 |
| chr4_-_62150810 | 13.30 |
ENSMUST00000077719.3
|
Mup21
|
major urinary protein 21 |
| chr7_+_13623967 | 13.18 |
ENSMUST00000108525.2
|
Sult2a5
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 5 |
| chr4_-_96664112 | 13.00 |
ENSMUST00000030299.7
|
Cyp2j5
|
cytochrome P450, family 2, subfamily j, polypeptide 5 |
| chr4_-_60222580 | 12.88 |
ENSMUST00000095058.4
ENSMUST00000163931.1 |
Mup8
|
major urinary protein 8 |
| chr7_+_51880312 | 12.79 |
ENSMUST00000145049.1
|
Gas2
|
growth arrest specific 2 |
| chr13_-_23914998 | 12.61 |
ENSMUST00000021769.8
ENSMUST00000110407.2 |
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr1_+_88070765 | 12.40 |
ENSMUST00000073772.4
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr8_+_105131800 | 11.66 |
ENSMUST00000161289.1
|
Ces4a
|
carboxylesterase 4A |
| chr1_-_150466165 | 11.12 |
ENSMUST00000162367.1
ENSMUST00000161611.1 ENSMUST00000161320.1 ENSMUST00000159035.1 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr10_+_128971191 | 10.77 |
ENSMUST00000181142.1
|
9030616G12Rik
|
RIKEN cDNA 9030616G12 gene |
| chr7_+_26307190 | 10.55 |
ENSMUST00000098657.3
|
Cyp2a4
|
cytochrome P450, family 2, subfamily a, polypeptide 4 |
| chr19_-_8405060 | 10.52 |
ENSMUST00000064507.5
ENSMUST00000120540.1 ENSMUST00000096269.4 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr14_+_41105359 | 10.18 |
ENSMUST00000047286.6
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr5_+_7960445 | 10.17 |
ENSMUST00000115421.1
|
Steap4
|
STEAP family member 4 |
| chr6_-_141946960 | 10.16 |
ENSMUST00000042119.5
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr19_-_40187277 | 9.96 |
ENSMUST00000051846.6
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr15_-_34495180 | 9.73 |
ENSMUST00000022946.5
|
Hrsp12
|
heat-responsive protein 12 |
| chr1_+_88166004 | 9.60 |
ENSMUST00000097659.4
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr1_-_180199587 | 9.40 |
ENSMUST00000161743.1
|
Adck3
|
aarF domain containing kinase 3 |
| chr4_-_108118504 | 9.38 |
ENSMUST00000149106.1
|
Scp2
|
sterol carrier protein 2, liver |
| chr3_-_67515487 | 9.37 |
ENSMUST00000178314.1
ENSMUST00000054825.4 |
Rarres1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chrX_+_59999436 | 9.30 |
ENSMUST00000033477.4
|
F9
|
coagulation factor IX |
| chr6_-_141946791 | 9.20 |
ENSMUST00000168119.1
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr2_+_122147680 | 9.15 |
ENSMUST00000102476.4
|
B2m
|
beta-2 microglobulin |
| chr7_+_67647405 | 8.93 |
ENSMUST00000032774.8
ENSMUST00000107471.1 |
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr17_-_12507704 | 8.73 |
ENSMUST00000024595.2
|
Slc22a3
|
solute carrier family 22 (organic cation transporter), member 3 |
| chr19_-_20727533 | 8.69 |
ENSMUST00000025656.3
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr9_+_78289923 | 8.60 |
ENSMUST00000119823.1
ENSMUST00000121273.1 |
Gm10639
|
predicted gene 10639 |
| chr15_+_4727265 | 8.56 |
ENSMUST00000162350.1
|
C6
|
complement component 6 |
| chr2_+_173153048 | 8.53 |
ENSMUST00000029017.5
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr7_+_119526269 | 8.45 |
ENSMUST00000066465.1
|
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr14_+_66635251 | 8.44 |
ENSMUST00000159365.1
ENSMUST00000054661.1 ENSMUST00000159068.1 |
Adra1a
|
adrenergic receptor, alpha 1a |
| chr15_+_9279829 | 8.39 |
ENSMUST00000022861.8
|
Ugt3a1
|
UDP glycosyltransferases 3 family, polypeptide A1 |
| chr2_+_43555321 | 8.34 |
ENSMUST00000028223.2
|
Kynu
|
kynureninase (L-kynurenine hydrolase) |
| chr2_-_86347764 | 8.30 |
ENSMUST00000099894.2
|
Olfr1055
|
olfactory receptor 1055 |
| chr8_-_93048192 | 8.23 |
ENSMUST00000095211.4
|
Ces1a
|
carboxylesterase 1A |
| chr10_+_21377290 | 8.15 |
ENSMUST00000042699.7
ENSMUST00000159163.1 |
Aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr19_-_8218832 | 8.08 |
ENSMUST00000113298.2
|
Slc22a29
|
solute carrier family 22. member 29 |
| chr19_-_39649046 | 8.00 |
ENSMUST00000067328.6
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr8_+_45069137 | 7.92 |
ENSMUST00000067984.7
|
Mtnr1a
|
melatonin receptor 1A |
| chr2_+_43555342 | 7.69 |
ENSMUST00000112826.1
ENSMUST00000050511.6 |
Kynu
|
kynureninase (L-kynurenine hydrolase) |
| chr4_-_108118528 | 7.64 |
ENSMUST00000030340.8
|
Scp2
|
sterol carrier protein 2, liver |
| chr6_-_85713205 | 7.49 |
ENSMUST00000160534.1
|
Gm4477
|
predicted gene 4477 |
| chr6_-_128526703 | 7.49 |
ENSMUST00000143664.1
ENSMUST00000112132.1 ENSMUST00000032510.7 |
Pzp
|
pregnancy zone protein |
| chr10_+_62071014 | 7.27 |
ENSMUST00000053865.5
|
Gm5424
|
predicted gene 5424 |
| chr19_+_26623419 | 7.24 |
ENSMUST00000176584.1
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_+_26808880 | 7.10 |
ENSMUST00000040944.7
|
Cyp2g1
|
cytochrome P450, family 2, subfamily g, polypeptide 1 |
| chr17_-_57228003 | 7.03 |
ENSMUST00000177046.1
ENSMUST00000024988.8 |
C3
|
complement component 3 |
| chr15_+_3270767 | 6.86 |
ENSMUST00000082424.4
ENSMUST00000159158.1 ENSMUST00000159216.1 ENSMUST00000160311.1 |
Sepp1
|
selenoprotein P, plasma, 1 |
| chr8_+_67490758 | 6.79 |
ENSMUST00000026677.3
|
Nat1
|
N-acetyl transferase 1 |
| chr18_+_87756280 | 6.79 |
ENSMUST00000091776.5
|
Gm5096
|
predicted gene 5096 |
| chr1_-_121332545 | 6.57 |
ENSMUST00000161068.1
|
Insig2
|
insulin induced gene 2 |
| chr3_+_60031754 | 6.56 |
ENSMUST00000029325.3
|
Aadac
|
arylacetamide deacetylase (esterase) |
| chr7_-_25477607 | 6.52 |
ENSMUST00000098669.1
ENSMUST00000098668.1 ENSMUST00000098666.2 |
Ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr15_-_76126538 | 6.38 |
ENSMUST00000054022.5
ENSMUST00000089654.3 |
BC024139
|
cDNA sequence BC024139 |
| chr12_+_8012359 | 6.30 |
ENSMUST00000171239.1
|
Apob
|
apolipoprotein B |
| chr4_+_42629719 | 6.26 |
ENSMUST00000166898.2
|
Gm2564
|
predicted gene 2564 |
| chr5_-_87482258 | 6.24 |
ENSMUST00000079811.6
ENSMUST00000144144.1 |
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr6_-_129237948 | 6.10 |
ENSMUST00000181238.1
ENSMUST00000180379.1 |
2310001H17Rik
|
RIKEN cDNA 2310001H17 gene |
| chr14_+_65971049 | 6.09 |
ENSMUST00000128539.1
|
Clu
|
clusterin |
| chr1_-_65179058 | 6.02 |
ENSMUST00000097709.4
|
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr15_-_3582596 | 6.00 |
ENSMUST00000161770.1
|
Ghr
|
growth hormone receptor |
| chr7_-_25539845 | 6.00 |
ENSMUST00000066503.7
ENSMUST00000064862.6 |
Ceacam2
|
carcinoembryonic antigen-related cell adhesion molecule 2 |
| chr13_+_4049001 | 5.97 |
ENSMUST00000118717.2
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr9_-_15301555 | 5.92 |
ENSMUST00000034414.8
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr5_-_91402905 | 5.91 |
ENSMUST00000121044.2
|
Btc
|
betacellulin, epidermal growth factor family member |
| chr3_+_89459118 | 5.86 |
ENSMUST00000029564.5
|
Pmvk
|
phosphomevalonate kinase |
| chr4_+_144893077 | 5.84 |
ENSMUST00000154208.1
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr10_+_63024512 | 5.82 |
ENSMUST00000020262.4
|
Pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chrX_-_143933089 | 5.77 |
ENSMUST00000087313.3
|
Dcx
|
doublecortin |
| chr10_+_62920630 | 5.76 |
ENSMUST00000044977.3
|
Slc25a16
|
solute carrier family 25 (mitochondrial carrier, Graves disease autoantigen), member 16 |
| chr18_-_38918642 | 5.75 |
ENSMUST00000040647.4
|
Fgf1
|
fibroblast growth factor 1 |
| chr15_+_4727202 | 5.64 |
ENSMUST00000161997.1
ENSMUST00000022788.8 |
C6
|
complement component 6 |
| chr8_-_93229517 | 5.62 |
ENSMUST00000176282.1
ENSMUST00000034173.7 |
Ces1e
|
carboxylesterase 1E |
| chr3_-_107986360 | 5.55 |
ENSMUST00000066530.6
|
Gstm2
|
glutathione S-transferase, mu 2 |
| chr7_+_27029074 | 5.54 |
ENSMUST00000075552.5
|
Cyp2a12
|
cytochrome P450, family 2, subfamily a, polypeptide 12 |
| chr5_-_116422858 | 5.34 |
ENSMUST00000036991.4
|
Hspb8
|
heat shock protein 8 |
| chr2_-_5676046 | 5.33 |
ENSMUST00000114987.3
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr19_-_7966000 | 5.30 |
ENSMUST00000182102.1
ENSMUST00000075619.4 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr3_+_94398517 | 5.24 |
ENSMUST00000050975.3
|
Lingo4
|
leucine rich repeat and Ig domain containing 4 |
| chr17_-_34862122 | 5.24 |
ENSMUST00000154526.1
|
Cfb
|
complement factor B |
| chr16_+_37580137 | 5.21 |
ENSMUST00000160847.1
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr11_+_115462464 | 5.20 |
ENSMUST00000106532.3
ENSMUST00000092445.5 ENSMUST00000153466.1 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
| chr3_+_133338936 | 5.18 |
ENSMUST00000150386.1
ENSMUST00000125858.1 |
Ppa2
|
pyrophosphatase (inorganic) 2 |
| chr5_+_135009152 | 5.14 |
ENSMUST00000111216.1
ENSMUST00000046999.8 |
Abhd11
|
abhydrolase domain containing 11 |
| chr9_+_77917364 | 5.11 |
ENSMUST00000034904.7
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
| chr1_-_121332571 | 5.08 |
ENSMUST00000071064.6
|
Insig2
|
insulin induced gene 2 |
| chr4_+_144892813 | 5.03 |
ENSMUST00000105744.1
ENSMUST00000171001.1 |
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr5_-_92328068 | 5.03 |
ENSMUST00000113093.3
|
Cxcl9
|
chemokine (C-X-C motif) ligand 9 |
| chrX_+_103321398 | 5.00 |
ENSMUST00000033689.2
|
Cdx4
|
caudal type homeobox 4 |
| chr5_+_90561102 | 4.98 |
ENSMUST00000094615.4
|
5830473C10Rik
|
RIKEN cDNA 5830473C10 gene |
| chr11_+_75468040 | 4.98 |
ENSMUST00000043598.7
ENSMUST00000108435.1 |
Tlcd2
|
TLC domain containing 2 |
| chr2_+_155517948 | 4.97 |
ENSMUST00000029135.8
ENSMUST00000065973.2 ENSMUST00000103142.5 |
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr9_+_119341487 | 4.94 |
ENSMUST00000175743.1
ENSMUST00000176397.1 |
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chr14_+_65970610 | 4.90 |
ENSMUST00000127387.1
|
Clu
|
clusterin |
| chr11_+_94211431 | 4.89 |
ENSMUST00000041589.5
|
Tob1
|
transducer of ErbB-2.1 |
| chr6_+_17463749 | 4.89 |
ENSMUST00000115443.1
|
Met
|
met proto-oncogene |
| chr5_-_137921612 | 4.89 |
ENSMUST00000031741.7
|
Cyp3a13
|
cytochrome P450, family 3, subfamily a, polypeptide 13 |
| chr17_-_34862473 | 4.88 |
ENSMUST00000025229.4
ENSMUST00000176203.2 ENSMUST00000128767.1 |
Cfb
|
complement factor B |
| chr1_+_21240581 | 4.87 |
ENSMUST00000027067.8
|
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr17_+_80127447 | 4.86 |
ENSMUST00000039205.4
|
Galm
|
galactose mutarotase |
| chr1_+_21240597 | 4.85 |
ENSMUST00000121676.1
|
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr8_+_94525067 | 4.84 |
ENSMUST00000098489.4
|
Nlrc5
|
NLR family, CARD domain containing 5 |
| chr16_+_22951072 | 4.83 |
ENSMUST00000023590.8
|
Hrg
|
histidine-rich glycoprotein |
| chr5_-_147894804 | 4.80 |
ENSMUST00000118527.1
ENSMUST00000031655.3 ENSMUST00000138244.1 |
Slc46a3
|
solute carrier family 46, member 3 |
| chr8_+_104591464 | 4.76 |
ENSMUST00000059588.6
|
Pdp2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr6_+_54269135 | 4.74 |
ENSMUST00000114401.1
|
Chn2
|
chimerin (chimaerin) 2 |
| chr11_-_9039585 | 4.74 |
ENSMUST00000043377.5
|
Sun3
|
Sad1 and UNC84 domain containing 3 |
| chr8_-_110805863 | 4.73 |
ENSMUST00000150680.1
ENSMUST00000076846.4 |
Il34
|
interleukin 34 |
| chr5_-_86906937 | 4.69 |
ENSMUST00000031181.9
ENSMUST00000113333.1 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr7_-_13989588 | 4.69 |
ENSMUST00000165167.1
ENSMUST00000108520.2 |
Sult2a4
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 4 |
| chr3_-_63964659 | 4.67 |
ENSMUST00000161659.1
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr10_-_24836165 | 4.65 |
ENSMUST00000020169.7
|
Enpp3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr1_+_88095054 | 4.63 |
ENSMUST00000150634.1
ENSMUST00000058237.7 |
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C |
| chr10_+_62920648 | 4.62 |
ENSMUST00000144459.1
|
Slc25a16
|
solute carrier family 25 (mitochondrial carrier, Graves disease autoantigen), member 16 |
| chr13_-_19307551 | 4.60 |
ENSMUST00000103561.1
|
Tcrg-C2
|
T-cell receptor gamma, constant 2 |
| chr7_-_72306595 | 4.58 |
ENSMUST00000079323.5
|
Mctp2
|
multiple C2 domains, transmembrane 2 |
| chr3_+_19985612 | 4.57 |
ENSMUST00000172860.1
|
Cp
|
ceruloplasmin |
| chr8_-_45358737 | 4.53 |
ENSMUST00000155230.1
ENSMUST00000135912.1 |
Fam149a
|
family with sequence similarity 149, member A |
| chr11_-_69369377 | 4.49 |
ENSMUST00000092971.6
ENSMUST00000108661.1 |
Chd3
|
chromodomain helicase DNA binding protein 3 |
| chr8_+_46010596 | 4.44 |
ENSMUST00000110381.2
|
Lrp2bp
|
Lrp2 binding protein |
| chr14_+_65971164 | 4.44 |
ENSMUST00000144619.1
|
Clu
|
clusterin |
| chr3_+_97158767 | 4.44 |
ENSMUST00000090759.4
|
Acp6
|
acid phosphatase 6, lysophosphatidic |
| chr1_+_88055377 | 4.43 |
ENSMUST00000138182.1
ENSMUST00000113142.3 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr9_-_44802951 | 4.43 |
ENSMUST00000044694.6
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr1_-_162866502 | 4.40 |
ENSMUST00000046049.7
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr19_+_18749983 | 4.38 |
ENSMUST00000040489.7
|
Trpm6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr8_-_109579056 | 4.37 |
ENSMUST00000074898.6
|
Hp
|
haptoglobin |
| chr9_-_65908676 | 4.37 |
ENSMUST00000119245.1
ENSMUST00000134338.1 ENSMUST00000179395.1 |
Trip4
|
thyroid hormone receptor interactor 4 |
| chr3_-_95904683 | 4.35 |
ENSMUST00000147962.1
ENSMUST00000036181.8 |
Car14
|
carbonic anhydrase 14 |
| chr2_-_148040196 | 4.35 |
ENSMUST00000136555.1
|
9030622O22Rik
|
RIKEN cDNA 9030622O22 gene |
| chr17_+_36958571 | 4.33 |
ENSMUST00000040177.6
|
Znrd1as
|
Znrd1 antisense |
| chr13_+_4059565 | 4.32 |
ENSMUST00000041768.6
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr3_+_60006743 | 4.31 |
ENSMUST00000169794.1
|
Aadacl2
|
arylacetamide deacetylase-like 2 |
| chr4_+_144893127 | 4.24 |
ENSMUST00000142808.1
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr17_+_36958623 | 4.23 |
ENSMUST00000173814.1
|
Znrd1as
|
Znrd1 antisense |
| chr17_+_35470083 | 4.22 |
ENSMUST00000174525.1
ENSMUST00000068291.6 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr18_-_35498856 | 4.18 |
ENSMUST00000025215.8
|
Sil1
|
endoplasmic reticulum chaperone SIL1 homolog (S. cerevisiae) |
| chr6_-_124888192 | 4.18 |
ENSMUST00000024044.6
|
Cd4
|
CD4 antigen |
| chr10_-_128589650 | 4.18 |
ENSMUST00000082059.6
|
Erbb3
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) |
| chr1_+_171214013 | 4.17 |
ENSMUST00000111328.1
|
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr1_+_88055467 | 4.16 |
ENSMUST00000173325.1
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr10_-_24101951 | 4.16 |
ENSMUST00000170267.1
|
Taar8c
|
trace amine-associated receptor 8C |
| chr6_+_90465287 | 4.16 |
ENSMUST00000113530.1
|
Klf15
|
Kruppel-like factor 15 |
| chr7_+_67655414 | 4.15 |
ENSMUST00000107470.1
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr4_-_155361322 | 4.13 |
ENSMUST00000105624.1
|
Prkcz
|
protein kinase C, zeta |
| chr18_+_20944607 | 4.10 |
ENSMUST00000050004.1
|
Rnf125
|
ring finger protein 125 |
| chr3_-_82903963 | 4.10 |
ENSMUST00000029632.6
|
Lrat
|
lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) |
| chr14_-_45477856 | 4.09 |
ENSMUST00000141424.1
|
Fermt2
|
fermitin family homolog 2 (Drosophila) |
| chr8_-_41215146 | 4.08 |
ENSMUST00000034003.4
|
Fgl1
|
fibrinogen-like protein 1 |
| chr18_-_84681966 | 4.08 |
ENSMUST00000168419.1
|
Cndp2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr13_+_55714624 | 4.07 |
ENSMUST00000021959.9
|
Txndc15
|
thioredoxin domain containing 15 |
| chr1_+_88138364 | 4.05 |
ENSMUST00000014263.4
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr17_+_79614900 | 4.03 |
ENSMUST00000040368.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr2_+_116900152 | 4.02 |
ENSMUST00000126467.1
ENSMUST00000128305.1 ENSMUST00000155323.1 |
D330050G23Rik
|
RIKEN cDNA D330050G23 gene |
| chr6_-_85765744 | 4.00 |
ENSMUST00000050780.7
|
Cml3
|
camello-like 3 |
| chr1_-_140183283 | 3.99 |
ENSMUST00000111977.1
|
Cfh
|
complement component factor h |
| chr1_-_140183404 | 3.97 |
ENSMUST00000066859.6
ENSMUST00000111976.2 |
Cfh
|
complement component factor h |
| chr3_+_19957088 | 3.97 |
ENSMUST00000108328.1
|
Cp
|
ceruloplasmin |
| chr3_+_19957037 | 3.92 |
ENSMUST00000091309.5
ENSMUST00000108329.1 ENSMUST00000003714.6 |
Cp
|
ceruloplasmin |
| chr4_-_108031938 | 3.90 |
ENSMUST00000106708.1
|
Podn
|
podocan |
| chr6_+_149141513 | 3.88 |
ENSMUST00000047531.9
ENSMUST00000179873.1 ENSMUST00000111548.1 ENSMUST00000111547.1 ENSMUST00000134306.1 |
Mettl20
|
methyltransferase like 20 |
| chr7_-_127273919 | 3.87 |
ENSMUST00000082428.3
|
Sephs2
|
selenophosphate synthetase 2 |
| chr6_+_68161415 | 3.84 |
ENSMUST00000168090.1
|
Igkv1-115
|
immunoglobulin kappa variable 1-115 |
| chr5_+_29195983 | 3.82 |
ENSMUST00000160888.1
ENSMUST00000159272.1 ENSMUST00000001247.5 ENSMUST00000161398.1 ENSMUST00000160246.1 |
Rnf32
|
ring finger protein 32 |
| chr11_+_102041509 | 3.82 |
ENSMUST00000123895.1
ENSMUST00000017453.5 ENSMUST00000107163.2 ENSMUST00000107164.2 |
Cd300lg
|
CD300 antigen like family member G |
| chr9_+_74976096 | 3.79 |
ENSMUST00000081746.5
|
Fam214a
|
family with sequence similarity 214, member A |
| chr8_+_45999297 | 3.77 |
ENSMUST00000110380.1
ENSMUST00000066451.3 |
Lrp2bp
|
Lrp2 binding protein |
| chr2_+_25653111 | 3.77 |
ENSMUST00000038482.6
|
Lcn8
|
lipocalin 8 |
| chr14_+_52810934 | 3.76 |
ENSMUST00000103646.3
|
Trav10d
|
T cell receptor alpha variable 10D |
| chr7_+_4237699 | 3.74 |
ENSMUST00000117550.1
|
Lilra5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| chr9_-_71168657 | 3.73 |
ENSMUST00000113570.1
|
Aqp9
|
aquaporin 9 |
| chr12_-_81485073 | 3.73 |
ENSMUST00000166723.1
ENSMUST00000110340.2 ENSMUST00000168463.1 ENSMUST00000169124.1 ENSMUST00000002757.4 |
Cox16
|
cytochrome c oxidase assembly protein 16 |
| chr17_-_45686899 | 3.72 |
ENSMUST00000156254.1
|
Tmem63b
|
transmembrane protein 63b |
| chr16_+_42907563 | 3.70 |
ENSMUST00000151244.1
ENSMUST00000114694.2 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cebpe
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 16.0 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 4.7 | 47.3 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 4.6 | 77.7 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 4.4 | 146.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 4.3 | 13.0 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 4.3 | 17.0 | GO:0032385 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 4.2 | 21.2 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 3.9 | 54.4 | GO:0015747 | urate transport(GO:0015747) |
| 3.2 | 9.7 | GO:0043385 | mycotoxin metabolic process(GO:0043385) mycotoxin catabolic process(GO:0043387) aflatoxin metabolic process(GO:0046222) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound metabolic process(GO:1901376) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 3.0 | 78.5 | GO:0080184 | response to phenylpropanoid(GO:0080184) |
| 2.8 | 8.5 | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 2.8 | 8.4 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 2.8 | 11.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 2.8 | 13.8 | GO:0002477 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 2.6 | 15.4 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 2.5 | 10.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 2.2 | 6.5 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 2.0 | 6.0 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 2.0 | 6.0 | GO:0070342 | brown fat cell proliferation(GO:0070342) regulation of brown fat cell proliferation(GO:0070347) |
| 1.6 | 4.8 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 1.5 | 9.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 1.5 | 6.0 | GO:0001887 | selenium compound metabolic process(GO:0001887) selenocysteine metabolic process(GO:0016259) |
| 1.5 | 2.9 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 1.5 | 4.4 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 1.4 | 4.2 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 1.4 | 11.1 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 1.3 | 18.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.2 | 7.4 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 1.2 | 5.9 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 1.2 | 8.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.2 | 3.5 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 1.2 | 9.2 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 1.1 | 4.4 | GO:2000562 | regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 1.1 | 8.7 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 1.1 | 3.3 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 1.1 | 3.2 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 1.0 | 5.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 1.0 | 10.2 | GO:0033216 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 1.0 | 3.0 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 1.0 | 4.9 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 1.0 | 4.9 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 1.0 | 2.9 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.9 | 6.6 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.9 | 15.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.9 | 6.6 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.9 | 2.8 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.9 | 2.6 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.8 | 2.5 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.8 | 3.3 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.8 | 2.5 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.8 | 7.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.8 | 4.8 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.8 | 13.4 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.8 | 2.4 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.8 | 6.9 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.8 | 3.8 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.8 | 2.3 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.7 | 2.2 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.7 | 1.4 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.7 | 2.1 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.7 | 2.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.7 | 2.0 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.7 | 2.0 | GO:1904414 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.7 | 2.0 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.7 | 2.6 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.6 | 2.6 | GO:0046073 | dTMP biosynthetic process(GO:0006231) dTMP metabolic process(GO:0046073) |
| 0.6 | 3.2 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) |
| 0.6 | 3.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.6 | 1.9 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.6 | 3.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.6 | 1.8 | GO:1900369 | negative regulation of RNA interference(GO:1900369) regulation of histone demethylase activity (H3-K4 specific)(GO:1904173) |
| 0.6 | 2.4 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.6 | 16.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.6 | 6.0 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.6 | 2.4 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.6 | 4.1 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.6 | 4.7 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.6 | 3.5 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.6 | 2.9 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.6 | 10.8 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.6 | 7.7 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.6 | 3.3 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.6 | 3.3 | GO:0002786 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 0.5 | 8.7 | GO:0051608 | histamine transport(GO:0051608) |
| 0.5 | 1.1 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.5 | 2.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.5 | 5.4 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.5 | 2.7 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.5 | 14.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.5 | 9.0 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.5 | 1.6 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.5 | 4.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.5 | 4.7 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.5 | 4.6 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.5 | 1.5 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.5 | 4.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.5 | 4.1 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.5 | 2.0 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.5 | 2.5 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.5 | 2.5 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.5 | 5.9 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.5 | 6.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.5 | 2.4 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.5 | 4.4 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.5 | 1.9 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.5 | 0.5 | GO:2000137 | negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 0.5 | 1.4 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) cellular response to fructose stimulus(GO:0071332) |
| 0.5 | 5.9 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.5 | 1.4 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 0.4 | 4.9 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) |
| 0.4 | 3.1 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) |
| 0.4 | 4.4 | GO:0006983 | ER overload response(GO:0006983) |
| 0.4 | 2.6 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.4 | 2.2 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.4 | 1.3 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.4 | 1.3 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.4 | 2.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.4 | 1.6 | GO:0007227 | signal transduction downstream of smoothened(GO:0007227) positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.4 | 1.6 | GO:0014056 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) |
| 0.4 | 1.2 | GO:0090155 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.4 | 3.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.4 | 2.0 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.4 | 0.8 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.4 | 1.6 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.4 | 6.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.4 | 2.7 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.4 | 2.3 | GO:2001184 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.4 | 2.7 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.4 | 2.7 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.4 | 2.6 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.4 | 0.7 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.4 | 4.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.4 | 0.7 | GO:0006244 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.4 | 1.8 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.4 | 2.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.3 | 1.0 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.3 | 1.4 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.3 | 4.7 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.3 | 10.1 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 | 2.3 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.3 | 2.0 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.3 | 4.7 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.3 | 1.3 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.3 | 1.6 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.3 | 1.0 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.3 | 1.0 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.3 | 4.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.3 | 1.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.3 | 4.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.3 | 6.3 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.3 | 1.9 | GO:0044597 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.3 | 1.8 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.3 | 1.8 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.3 | 0.9 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.3 | 1.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.3 | 2.4 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.3 | 1.5 | GO:0009597 | detection of virus(GO:0009597) |
| 0.3 | 0.9 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.3 | 0.9 | GO:1900625 | positive regulation of mast cell cytokine production(GO:0032765) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.3 | 1.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.3 | 2.8 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.3 | 1.4 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.3 | 2.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.3 | 1.9 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.3 | 3.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.3 | 2.4 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.3 | 2.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.3 | 11.0 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.3 | 1.0 | GO:0034238 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) |
| 0.3 | 1.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.3 | 0.8 | GO:0036395 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.3 | 3.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.3 | 1.0 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.3 | 1.5 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.3 | 1.0 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.2 | 9.1 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.2 | 2.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 4.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.2 | 2.9 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 1.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.2 | 2.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 0.7 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.2 | 3.0 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 1.8 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.2 | 1.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.2 | 0.2 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.2 | 6.1 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.2 | 2.7 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 0.7 | GO:1903294 | regulation of glutamate secretion, neurotransmission(GO:1903294) positive regulation of glutamate secretion, neurotransmission(GO:1903296) |
| 0.2 | 0.7 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.2 | 0.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 4.4 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.2 | 0.4 | GO:1904348 | negative regulation of gastro-intestinal system smooth muscle contraction(GO:1904305) negative regulation of small intestine smooth muscle contraction(GO:1904348) |
| 0.2 | 0.7 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.2 | 4.6 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 0.2 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.2 | 2.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 1.3 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.2 | 0.8 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.2 | 1.0 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.2 | 1.9 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.2 | 2.7 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 0.8 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.2 | 1.4 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.2 | 5.5 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.2 | 0.4 | GO:0033864 | positive regulation of NAD(P)H oxidase activity(GO:0033864) |
| 0.2 | 2.6 | GO:0035337 | fatty-acyl-CoA metabolic process(GO:0035337) |
| 0.2 | 2.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 0.8 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.2 | 1.1 | GO:0018158 | protein oxidation(GO:0018158) |
| 0.2 | 1.5 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.2 | 3.1 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 | 0.7 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.2 | 0.5 | GO:0001878 | response to yeast(GO:0001878) |
| 0.2 | 0.9 | GO:0046110 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
| 0.2 | 0.7 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.2 | 2.1 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 1.2 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.2 | 2.6 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.2 | 1.4 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.2 | 2.9 | GO:0097688 | glutamate receptor clustering(GO:0097688) |
| 0.2 | 1.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.2 | 2.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 3.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.2 | 1.0 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.2 | 3.0 | GO:0006415 | translational termination(GO:0006415) |
| 0.2 | 1.3 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.2 | 2.6 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.2 | 7.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.2 | 1.4 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.2 | 1.9 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.2 | 0.6 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.2 | 0.8 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.2 | 7.6 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.2 | 0.8 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.2 | 0.9 | GO:0060526 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.1 | 0.3 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 3.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 18.6 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 1.6 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 1.4 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.7 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.1 | 0.4 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.1 | 2.7 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.4 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.7 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 2.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 2.0 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 4.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 1.0 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.1 | 0.5 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.4 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.1 | 1.9 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.1 | 0.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.1 | 0.4 | GO:0042697 | menopause(GO:0042697) |
| 0.1 | 1.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.4 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.1 | 0.5 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.1 | 1.0 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 0.1 | 0.9 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 1.4 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 1.6 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.1 | 0.9 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 0.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.9 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 2.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 2.0 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 1.2 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.1 | 5.9 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 0.6 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.1 | 0.3 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.1 | 3.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 1.5 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.8 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.1 | 1.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 19.5 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 3.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 5.2 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 2.1 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 0.9 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.1 | 0.6 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 0.9 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.4 | GO:0061356 | adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) cellular response to isoquinoline alkaloid(GO:0071317) |
| 0.1 | 1.0 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.6 | GO:0072017 | distal tubule development(GO:0072017) |
| 0.1 | 3.2 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 0.9 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.3 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.1 | 0.8 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.3 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.1 | 0.6 | GO:0051189 | molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 0.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 1.3 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.1 | 0.3 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 13.9 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.1 | 0.6 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 1.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.2 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.1 | 10.2 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 0.3 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 1.0 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.1 | 4.8 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.7 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.1 | 0.3 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 3.3 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.1 | 6.0 | GO:0048041 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.1 | 0.2 | GO:0046709 | ADP catabolic process(GO:0046032) IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.1 | 0.2 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.1 | 0.4 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.1 | 0.9 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 5.0 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 2.6 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.1 | 8.4 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 1.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.7 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 1.6 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 2.0 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 2.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.5 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 1.2 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 0.6 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.4 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 1.7 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 | 0.4 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 0.8 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 1.1 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.7 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.9 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.1 | 0.4 | GO:0051458 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.1 | 0.6 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.1 | 1.8 | GO:0007584 | response to nutrient(GO:0007584) |
| 0.1 | 0.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 2.2 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 | 2.1 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.2 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 0.8 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.2 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 0.8 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 0.2 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) |
| 0.1 | 17.2 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.6 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 1.3 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 1.0 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.6 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.3 | GO:0009191 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 2.2 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 1.3 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 8.6 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.4 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.3 | GO:0031053 | primary miRNA processing(GO:0031053) G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.1 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.9 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 0.7 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 1.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) sebaceous gland development(GO:0048733) |
| 0.0 | 1.6 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 2.3 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.6 | GO:0007595 | lactation(GO:0007595) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.8 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.2 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.7 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.0 | 0.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.4 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.7 | GO:0032835 | glomerulus development(GO:0032835) |
| 0.0 | 0.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.1 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 1.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.1 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 1.4 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.1 | GO:0002863 | positive regulation of inflammatory response to antigenic stimulus(GO:0002863) |
| 0.0 | 0.2 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.6 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.0 | 0.1 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.5 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.4 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.0 | 0.4 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.3 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 2.3 | GO:0030832 | regulation of actin filament length(GO:0030832) |
| 0.0 | 0.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.6 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.3 | GO:0060443 | mammary gland morphogenesis(GO:0060443) |
| 0.0 | 0.6 | GO:0006757 | ATP generation from ADP(GO:0006757) |
| 0.0 | 0.2 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.2 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.0 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.0 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.6 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 2.0 | 6.0 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 1.6 | 4.8 | GO:0032010 | phagolysosome(GO:0032010) |
| 1.5 | 11.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 1.3 | 17.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 1.3 | 14.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.3 | 5.1 | GO:0097447 | dendritic tree(GO:0097447) |
| 1.2 | 15.4 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 1.1 | 13.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 1.0 | 10.7 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.9 | 4.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.8 | 2.5 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.8 | 2.5 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.8 | 3.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.8 | 2.5 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.8 | 6.6 | GO:0042825 | TAP complex(GO:0042825) |
| 0.8 | 23.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.8 | 2.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.7 | 9.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.6 | 2.3 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.5 | 2.7 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.5 | 3.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.5 | 3.6 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.5 | 3.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.5 | 1.8 | GO:0071942 | XPC complex(GO:0071942) |
| 0.4 | 1.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.4 | 8.7 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.4 | 4.7 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.4 | 21.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.4 | 1.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.4 | 7.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.4 | 2.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.3 | 1.7 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.3 | 1.3 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 3.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.3 | 2.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 1.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.3 | 6.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.3 | 3.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.3 | 9.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.3 | 1.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.3 | 2.8 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.3 | 4.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.3 | 2.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.3 | 2.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.3 | 1.3 | GO:1990923 | PET complex(GO:1990923) |
| 0.2 | 11.2 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 1.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 7.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 0.7 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 2.6 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.2 | 0.9 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.2 | 3.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 2.5 | GO:0048500 | signal recognition particle(GO:0048500) |
| 0.2 | 0.9 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 1.4 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.2 | 2.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.2 | 1.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 84.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.2 | 2.0 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 37.8 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 4.5 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 1.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 2.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.2 | 4.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 2.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.2 | 0.5 | GO:0060473 | cortical granule(GO:0060473) |
| 0.2 | 0.8 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.2 | 0.8 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.2 | 2.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 1.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 1.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 2.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.4 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.7 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 1.7 | GO:0031254 | uropod(GO:0001931) flotillin complex(GO:0016600) cell trailing edge(GO:0031254) |
| 0.1 | 4.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.0 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 38.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 3.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 0.7 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 5.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 2.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.7 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 6.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 0.7 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 2.0 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 7.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.0 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.1 | 0.4 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.9 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 2.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 2.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 1.0 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.1 | 0.4 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 4.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 128.9 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.1 | 1.9 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 1.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.9 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 3.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 2.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 1.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.3 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.9 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 12.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 0.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 1.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 7.3 | GO:0005740 | mitochondrial envelope(GO:0005740) |
| 0.0 | 3.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 1.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 6.6 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.7 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 2.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 2.0 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.3 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.4 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 2.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 35.0 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 2.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 3.8 | GO:0098562 | cytoplasmic side of plasma membrane(GO:0009898) cytoplasmic side of membrane(GO:0098562) |
| 0.0 | 2.4 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 1.0 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 1.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 25.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.8 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.0 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.1 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 3.6 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.7 | 88.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 10.6 | 31.9 | GO:0005186 | insulin-activated receptor activity(GO:0005009) pheromone activity(GO:0005186) |
| 4.8 | 182.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 4.3 | 17.0 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 4.0 | 16.0 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 3.7 | 22.0 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 3.6 | 54.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 3.1 | 22.0 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 2.9 | 8.7 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 2.3 | 13.8 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 2.2 | 15.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 2.2 | 10.8 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 2.1 | 8.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 2.0 | 6.0 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 2.0 | 6.0 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 1.9 | 58.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 1.7 | 6.8 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 1.7 | 6.6 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 1.7 | 5.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 1.6 | 4.8 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 1.6 | 4.7 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 1.6 | 32.8 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 1.5 | 15.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.5 | 11.9 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 1.5 | 8.7 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 1.3 | 6.6 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 1.3 | 5.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 1.3 | 14.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 1.3 | 7.7 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 1.3 | 10.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 1.2 | 7.4 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) palmitoyl-CoA oxidase activity(GO:0016401) C-acetyltransferase activity(GO:0016453) |
| 1.2 | 4.9 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 1.2 | 4.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 1.2 | 8.2 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 1.1 | 17.9 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) aromatase activity(GO:0070330) |
| 1.1 | 3.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 1.0 | 5.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.0 | 19.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.9 | 4.5 | GO:0015205 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.9 | 2.7 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.9 | 3.6 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.9 | 3.5 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.9 | 2.6 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) serine binding(GO:0070905) |
| 0.8 | 2.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.8 | 2.4 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.8 | 2.4 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.8 | 2.4 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.8 | 3.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.8 | 4.7 | GO:0035529 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 0.8 | 7.6 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.7 | 24.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.7 | 6.7 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.7 | 2.2 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.7 | 7.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.7 | 10.7 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.7 | 2.7 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.7 | 3.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.6 | 15.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.6 | 4.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.6 | 2.4 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.6 | 4.8 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.6 | 4.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.6 | 3.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.6 | 2.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.6 | 27.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.5 | 2.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.5 | 2.5 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.5 | 2.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.5 | 1.5 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.5 | 5.9 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.5 | 8.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.5 | 2.8 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.5 | 1.9 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.5 | 6.8 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.4 | 3.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.4 | 4.4 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.4 | 12.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.4 | 6.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.4 | 1.3 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.4 | 4.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 0.4 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.4 | 3.6 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.4 | 1.2 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.4 | 2.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.4 | 1.6 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.4 | 22.3 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.4 | 4.2 | GO:0046977 | TAP binding(GO:0046977) |
| 0.4 | 3.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.4 | 1.9 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.4 | 3.0 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.4 | 8.8 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.4 | 6.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.4 | 5.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 1.4 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.4 | 1.4 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.4 | 3.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.4 | 2.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.4 | 2.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.4 | 1.4 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.3 | 3.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.3 | 4.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.3 | 4.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.3 | 2.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 10.6 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.3 | 1.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.3 | 1.8 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.3 | 3.7 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.3 | 0.8 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.3 | 5.3 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.3 | 4.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 2.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.3 | 2.7 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.3 | 5.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.3 | 0.8 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.3 | 1.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.3 | 2.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 2.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.3 | 5.6 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.3 | 4.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 1.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 5.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 1.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.2 | 1.7 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.2 | 11.2 | GO:0043531 | ADP binding(GO:0043531) |
| 0.2 | 1.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.2 | 2.4 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.2 | 4.8 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.2 | 1.4 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.2 | 7.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.2 | 2.5 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.2 | 0.9 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.2 | 3.8 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.2 | 3.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.2 | 0.9 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.2 | 0.9 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.2 | 1.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 0.8 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.2 | 4.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 1.8 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.2 | 1.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 3.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 2.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 1.5 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.2 | 0.8 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 2.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 1.3 | GO:0008312 | signal recognition particle binding(GO:0005047) 7S RNA binding(GO:0008312) |
| 0.2 | 40.3 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.2 | 1.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 0.9 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.2 | 3.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.2 | 6.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.2 | 0.9 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 0.9 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.2 | 1.9 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 4.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 1.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 2.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 1.2 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 1.2 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 22.3 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.2 | 3.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 0.5 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.2 | 1.3 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.2 | 1.3 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.2 | 4.0 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.2 | 2.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 1.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.2 | 0.5 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.2 | 0.8 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 1.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 2.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.7 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 1.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 4.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.9 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.5 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 2.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 2.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.4 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.1 | 4.6 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 0.6 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 0.4 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.1 | 18.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 1.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 1.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.4 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 1.6 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 2.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 5.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 4.7 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 3.5 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 1.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 3.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 3.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 4.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 4.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 7.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.4 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 3.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.9 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.6 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 1.0 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.5 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.1 | 1.7 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.4 | GO:0004985 | melanocortin receptor activity(GO:0004977) opioid receptor activity(GO:0004985) |
| 0.1 | 0.8 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.6 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 2.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.0 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 1.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 12.7 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.1 | 2.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.7 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.4 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 2.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.8 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 2.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 4.7 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 1.0 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 2.2 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 1.0 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 2.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 2.9 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 1.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.1 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.7 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 1.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.6 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.9 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.1 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 2.4 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 1.2 | GO:0072341 | modified amino acid binding(GO:0072341) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 2.2 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.6 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 1.7 | GO:0016853 | isomerase activity(GO:0016853) |
| 0.0 | 1.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.3 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.9 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 1.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.1 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 3.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.5 | 10.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.5 | 8.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.4 | 2.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.4 | 11.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.4 | 18.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 7.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 0.7 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.2 | 17.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 9.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 10.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 4.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 10.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 13.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 5.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.2 | 10.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.2 | 3.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 12.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 5.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 11.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 10.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 6.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 9.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 5.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 5.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 6.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 1.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 4.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 0.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 0.8 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 1.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 0.7 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.9 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 2.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 2.2 | 28.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 1.8 | 25.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.3 | 17.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 1.0 | 4.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 1.0 | 22.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.9 | 16.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.9 | 16.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.8 | 5.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.7 | 10.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.7 | 9.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.6 | 3.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.6 | 40.8 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.5 | 15.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.5 | 9.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.5 | 6.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.4 | 11.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.4 | 8.0 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.4 | 4.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.4 | 4.7 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.4 | 15.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.4 | 9.6 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.3 | 6.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.3 | 5.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.3 | 3.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 8.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 6.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.3 | 5.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.3 | 20.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 11.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.3 | 4.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.3 | 9.8 | REACTOME BIOLOGICAL OXIDATIONS | Genes involved in Biological oxidations |
| 0.2 | 7.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.2 | 3.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 5.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.2 | 3.7 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 3.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 1.5 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.2 | 3.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 1.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 4.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.2 | 18.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 18.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 4.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 1.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 1.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 1.3 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.2 | 2.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 5.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 4.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 4.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 3.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 1.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 5.1 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 1.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.2 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 1.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.0 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 3.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 2.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.6 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.1 | 1.0 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 0.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 0.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.0 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 1.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.9 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.1 | 3.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 3.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 0.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.2 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 1.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.9 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.6 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.0 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 2.1 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 1.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.7 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 3.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.1 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |