Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for E2f8
Z-value: 1.49
Transcription factors associated with E2f8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f8
|
ENSMUSG00000046179.11 | E2F transcription factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f8 | mm10_v2_chr7_-_48881596_48881619 | 0.77 | 4.3e-08 | Click! |
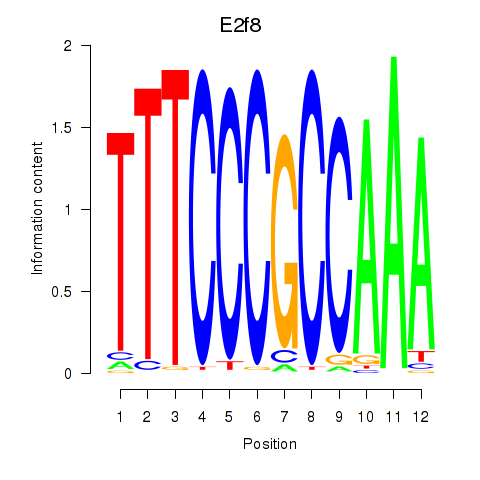
Activity profile of E2f8 motif
Sorted Z-values of E2f8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_90695706 | 18.19 |
ENSMUST00000069960.5
ENSMUST00000117167.1 |
S100a9
|
S100 calcium binding protein A9 (calgranulin B) |
| chr10_-_21160925 | 14.62 |
ENSMUST00000020158.6
|
Myb
|
myeloblastosis oncogene |
| chr12_+_109540979 | 12.59 |
ENSMUST00000129245.1
ENSMUST00000143836.1 ENSMUST00000124106.1 |
Meg3
|
maternally expressed 3 |
| chr11_+_98907801 | 11.59 |
ENSMUST00000092706.6
|
Cdc6
|
cell division cycle 6 |
| chr17_+_56303321 | 11.40 |
ENSMUST00000001258.8
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr2_+_163054682 | 11.39 |
ENSMUST00000018005.3
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr5_+_123749696 | 10.87 |
ENSMUST00000031366.7
|
Kntc1
|
kinetochore associated 1 |
| chr17_+_56303396 | 10.53 |
ENSMUST00000113038.1
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr8_+_75109528 | 10.18 |
ENSMUST00000164309.1
|
Mcm5
|
minichromosome maintenance deficient 5, cell division cycle 46 (S. cerevisiae) |
| chr12_+_24708984 | 9.21 |
ENSMUST00000154588.1
|
Rrm2
|
ribonucleotide reductase M2 |
| chr17_+_56040350 | 9.11 |
ENSMUST00000002914.8
|
Chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr6_-_88898664 | 8.41 |
ENSMUST00000058011.6
|
Mcm2
|
minichromosome maintenance deficient 2 mitotin (S. cerevisiae) |
| chr16_-_18811615 | 8.23 |
ENSMUST00000096990.3
|
Cdc45
|
cell division cycle 45 |
| chr12_+_24708241 | 7.70 |
ENSMUST00000020980.5
|
Rrm2
|
ribonucleotide reductase M2 |
| chr1_-_128359610 | 7.28 |
ENSMUST00000027601.4
|
Mcm6
|
minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) |
| chr9_-_36726374 | 7.08 |
ENSMUST00000172702.2
ENSMUST00000172742.1 ENSMUST00000034625.5 |
Chek1
|
checkpoint kinase 1 |
| chr10_+_128015157 | 6.22 |
ENSMUST00000178041.1
ENSMUST00000026461.7 |
Prim1
|
DNA primase, p49 subunit |
| chr2_-_34913976 | 5.86 |
ENSMUST00000028232.3
|
Phf19
|
PHD finger protein 19 |
| chr8_-_107065632 | 5.35 |
ENSMUST00000034393.5
|
Tmed6
|
transmembrane emp24 protein transport domain containing 6 |
| chr16_-_15637277 | 5.25 |
ENSMUST00000023353.3
|
Mcm4
|
minichromosome maintenance deficient 4 homolog (S. cerevisiae) |
| chr19_+_38931008 | 4.89 |
ENSMUST00000145051.1
|
Hells
|
helicase, lymphoid specific |
| chr13_+_19623163 | 4.85 |
ENSMUST00000002883.5
|
Sfrp4
|
secreted frizzled-related protein 4 |
| chr15_+_61985540 | 4.43 |
ENSMUST00000159327.1
ENSMUST00000167731.1 |
Myc
|
myelocytomatosis oncogene |
| chr11_-_101785252 | 4.41 |
ENSMUST00000164750.1
ENSMUST00000107176.1 ENSMUST00000017868.6 |
Etv4
|
ets variant gene 4 (E1A enhancer binding protein, E1AF) |
| chr15_+_61985377 | 4.32 |
ENSMUST00000161976.1
ENSMUST00000022971.7 |
Myc
|
myelocytomatosis oncogene |
| chr7_-_44548733 | 4.28 |
ENSMUST00000145956.1
ENSMUST00000049343.8 |
Pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr12_+_110279228 | 3.92 |
ENSMUST00000097228.4
|
Dio3
|
deiodinase, iodothyronine type III |
| chr6_+_35177610 | 3.84 |
ENSMUST00000170234.1
|
Nup205
|
nucleoporin 205 |
| chr17_+_8525369 | 3.83 |
ENSMUST00000115715.1
|
Pde10a
|
phosphodiesterase 10A |
| chr19_+_38930909 | 3.81 |
ENSMUST00000025965.5
|
Hells
|
helicase, lymphoid specific |
| chr6_+_35177386 | 3.45 |
ENSMUST00000043815.9
|
Nup205
|
nucleoporin 205 |
| chr7_-_115824699 | 3.23 |
ENSMUST00000169129.1
|
Sox6
|
SRY-box containing gene 6 |
| chr7_+_102065713 | 3.21 |
ENSMUST00000094129.2
ENSMUST00000094130.2 ENSMUST00000084843.3 |
Trpc2
|
transient receptor potential cation channel, subfamily C, member 2 |
| chr3_-_126998408 | 3.19 |
ENSMUST00000182764.1
ENSMUST00000044443.8 |
Ank2
|
ankyrin 2, brain |
| chr2_-_45113255 | 3.13 |
ENSMUST00000068415.4
ENSMUST00000127520.1 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr7_+_110122299 | 3.12 |
ENSMUST00000033326.8
|
Wee1
|
WEE 1 homolog 1 (S. pombe) |
| chr4_+_132768325 | 3.04 |
ENSMUST00000102561.4
|
Rpa2
|
replication protein A2 |
| chr9_+_103305156 | 2.99 |
ENSMUST00000035164.3
|
Topbp1
|
topoisomerase (DNA) II binding protein 1 |
| chr10_+_42860348 | 2.92 |
ENSMUST00000063063.7
|
Scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr5_-_33652296 | 2.86 |
ENSMUST00000151081.1
ENSMUST00000101354.3 |
Slbp
|
stem-loop binding protein |
| chr2_-_154569845 | 2.78 |
ENSMUST00000103145.4
|
E2f1
|
E2F transcription factor 1 |
| chr7_+_102065485 | 2.72 |
ENSMUST00000106950.1
ENSMUST00000146450.1 |
Trpc2
|
transient receptor potential cation channel, subfamily C, member 2 |
| chr16_+_10835046 | 2.30 |
ENSMUST00000037913.8
|
Rmi2
|
RMI2, RecQ mediated genome instability 2, homolog (S. cerevisiae) |
| chrX_+_49470450 | 2.24 |
ENSMUST00000114904.3
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chrX_+_49470555 | 2.23 |
ENSMUST00000042444.6
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr5_-_33652339 | 2.22 |
ENSMUST00000075670.6
|
Slbp
|
stem-loop binding protein |
| chr2_-_45112890 | 2.08 |
ENSMUST00000076836.6
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr15_+_78926720 | 2.06 |
ENSMUST00000089377.5
|
Lgals1
|
lectin, galactose binding, soluble 1 |
| chr2_-_154569720 | 2.03 |
ENSMUST00000000894.5
|
E2f1
|
E2F transcription factor 1 |
| chr10_-_128525859 | 1.88 |
ENSMUST00000026427.6
|
Esyt1
|
extended synaptotagmin-like protein 1 |
| chr13_+_44731265 | 1.82 |
ENSMUST00000173246.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr7_-_48881596 | 1.70 |
ENSMUST00000119223.1
|
E2f8
|
E2F transcription factor 8 |
| chr17_+_34377124 | 1.58 |
ENSMUST00000080254.5
|
Btnl1
|
butyrophilin-like 1 |
| chr2_-_45113216 | 1.54 |
ENSMUST00000124942.1
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr13_-_66851513 | 1.45 |
ENSMUST00000169322.1
|
Gm17404
|
predicted gene, 17404 |
| chr5_+_129020069 | 1.45 |
ENSMUST00000031383.7
ENSMUST00000111343.1 |
Ran
|
RAN, member RAS oncogene family |
| chr13_+_44731281 | 1.25 |
ENSMUST00000174086.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr5_+_135187251 | 1.21 |
ENSMUST00000002825.5
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr5_-_65335564 | 1.21 |
ENSMUST00000172780.1
|
Rfc1
|
replication factor C (activator 1) 1 |
| chr2_-_157204483 | 1.18 |
ENSMUST00000029170.7
|
Rbl1
|
retinoblastoma-like 1 (p107) |
| chr15_+_101174096 | 1.18 |
ENSMUST00000000544.9
|
Acvr1b
|
activin A receptor, type 1B |
| chr6_-_125286015 | 1.07 |
ENSMUST00000088246.5
|
Tuba3a
|
tubulin, alpha 3A |
| chr16_+_15637844 | 0.98 |
ENSMUST00000023352.8
|
Prkdc
|
protein kinase, DNA activated, catalytic polypeptide |
| chr17_+_33909481 | 0.95 |
ENSMUST00000173626.1
ENSMUST00000174541.1 |
Daxx
|
Fas death domain-associated protein |
| chr5_+_111581422 | 0.92 |
ENSMUST00000064930.3
|
C130026L21Rik
|
RIKEN cDNA C130026L21 gene |
| chr15_+_55557575 | 0.87 |
ENSMUST00000170046.1
|
Mtbp
|
Mdm2, transformed 3T3 cell double minute p53 binding protein |
| chr5_+_150673739 | 0.86 |
ENSMUST00000016569.4
ENSMUST00000038900.8 |
Pds5b
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chrX_+_153832225 | 0.85 |
ENSMUST00000148708.1
ENSMUST00000123264.1 ENSMUST00000049999.8 |
Spin2c
|
spindlin family, member 2C |
| chr11_+_88047693 | 0.81 |
ENSMUST00000079866.4
|
Srsf1
|
serine/arginine-rich splicing factor 1 |
| chr11_+_26387194 | 0.80 |
ENSMUST00000109509.1
ENSMUST00000136830.1 |
Fancl
|
Fanconi anemia, complementation group L |
| chr15_+_55557399 | 0.71 |
ENSMUST00000022998.7
|
Mtbp
|
Mdm2, transformed 3T3 cell double minute p53 binding protein |
| chr12_-_12941827 | 0.71 |
ENSMUST00000043396.7
|
Mycn
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian) |
| chr7_-_48881032 | 0.70 |
ENSMUST00000058745.8
|
E2f8
|
E2F transcription factor 8 |
| chr17_+_33909577 | 0.70 |
ENSMUST00000170075.2
|
Daxx
|
Fas death domain-associated protein |
| chr12_+_55836365 | 0.67 |
ENSMUST00000059250.6
|
Brms1l
|
breast cancer metastasis-suppressor 1-like |
| chr15_-_98093245 | 0.66 |
ENSMUST00000180657.1
|
Senp1
|
SUMO1/sentrin specific peptidase 1 |
| chr6_+_38551786 | 0.55 |
ENSMUST00000161227.1
|
Luc7l2
|
LUC7-like 2 (S. cerevisiae) |
| chr14_+_55222005 | 0.47 |
ENSMUST00000022820.5
|
Dhrs2
|
dehydrogenase/reductase member 2 |
| chr4_+_129189760 | 0.43 |
ENSMUST00000106054.2
ENSMUST00000001365.2 |
Yars
|
tyrosyl-tRNA synthetase |
| chr11_+_79993062 | 0.37 |
ENSMUST00000017692.8
ENSMUST00000163272.1 |
Suz12
|
suppressor of zeste 12 homolog (Drosophila) |
| chr1_-_168431896 | 0.34 |
ENSMUST00000176540.1
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr3_-_127553233 | 0.27 |
ENSMUST00000029588.5
|
Larp7
|
La ribonucleoprotein domain family, member 7 |
| chr13_-_66852017 | 0.25 |
ENSMUST00000059329.6
|
Gm17449
|
predicted gene, 17449 |
| chrX_-_41911877 | 0.25 |
ENSMUST00000047037.8
|
Thoc2
|
THO complex 2 |
| chr5_-_65335597 | 0.16 |
ENSMUST00000172660.1
ENSMUST00000172732.1 ENSMUST00000031092.8 |
Rfc1
|
replication factor C (activator 1) 1 |
| chr1_+_131910458 | 0.14 |
ENSMUST00000062264.6
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr8_+_40926220 | 0.13 |
ENSMUST00000034004.9
|
Pdgfrl
|
platelet-derived growth factor receptor-like |
| chr13_+_94875600 | 0.11 |
ENSMUST00000022195.10
|
Otp
|
orthopedia homolog (Drosophila) |
| chr4_-_129189600 | 0.09 |
ENSMUST00000117497.1
ENSMUST00000117350.1 |
S100pbp
|
S100P binding protein |
| chr17_+_33909409 | 0.08 |
ENSMUST00000173028.1
ENSMUST00000079421.7 |
Daxx
|
Fas death domain-associated protein |
| chr10_+_19356558 | 0.06 |
ENSMUST00000053225.5
|
Olig3
|
oligodendrocyte transcription factor 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 18.2 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 2.7 | 8.2 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 2.2 | 8.8 | GO:0090095 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 2.1 | 20.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 2.0 | 30.6 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 1.6 | 4.8 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 1.6 | 6.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 1.4 | 4.3 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 1.2 | 4.8 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 1.2 | 2.4 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 1.2 | 5.9 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 1.2 | 17.7 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 1.1 | 6.7 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 1.0 | 24.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.9 | 16.9 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.8 | 5.9 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.8 | 7.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.6 | 4.4 | GO:0060762 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.5 | 1.6 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.5 | 7.3 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.5 | 5.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.4 | 12.6 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.4 | 1.7 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.4 | 3.9 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.4 | 3.2 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.3 | 2.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.3 | 1.6 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.3 | 3.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.3 | 9.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 1.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.2 | 1.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 11.4 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.2 | 3.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 1.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.2 | 3.0 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.7 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 10.9 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.1 | 1.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.7 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 2.3 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.4 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.3 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 1.2 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:1990423 | RZZ complex(GO:1990423) |
| 3.4 | 16.9 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 3.0 | 9.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 2.7 | 8.2 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 2.3 | 11.4 | GO:0031523 | Myb complex(GO:0031523) |
| 1.9 | 28.2 | GO:0042555 | MCM complex(GO:0042555) |
| 1.5 | 7.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 1.4 | 4.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 1.2 | 6.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 1.0 | 4.8 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.7 | 5.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.5 | 34.6 | GO:0005657 | replication fork(GO:0005657) |
| 0.5 | 1.4 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.4 | 1.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.4 | 9.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 10.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.3 | 8.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.2 | 8.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.2 | 3.0 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.2 | 1.0 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 5.9 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 1.9 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.7 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 3.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.7 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.6 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 14.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 3.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 17.2 | GO:0005730 | nucleolus(GO:0005730) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 21.9 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 3.7 | 14.6 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 3.4 | 16.9 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 2.4 | 7.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 2.3 | 18.2 | GO:0050786 | Toll-like receptor 4 binding(GO:0035662) RAGE receptor binding(GO:0050786) |
| 1.9 | 26.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 1.7 | 5.1 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 1.3 | 3.9 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 1.3 | 11.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 1.1 | 9.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.7 | 2.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.5 | 5.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.4 | 3.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.3 | 1.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.3 | 12.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.3 | 4.3 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.3 | 7.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 1.4 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.2 | 3.0 | GO:0098505 | single-stranded telomeric DNA binding(GO:0043047) G-rich strand telomeric DNA binding(GO:0098505) |
| 0.2 | 0.7 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 4.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 1.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 6.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 8.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 6.2 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.1 | 1.4 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 6.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 3.1 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 1.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 4.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 8.7 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 3.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 7.2 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 3.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.7 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 4.5 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 10.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.6 | 57.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.5 | 18.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 11.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.3 | 14.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.2 | 8.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 1.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 2.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 5.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.7 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 4.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.4 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 2.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 39.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.9 | 33.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 1.1 | 10.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 1.0 | 13.5 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.7 | 11.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.4 | 5.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.3 | 8.7 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.3 | 8.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.2 | 3.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 14.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 3.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 1.0 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 4.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |