Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
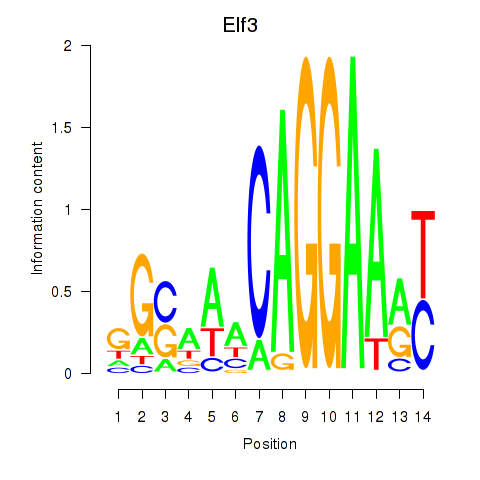
Results for Elf3
Z-value: 1.19
Transcription factors associated with Elf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Elf3
|
ENSMUSG00000003051.7 | E74-like factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Elf3 | mm10_v2_chr1_-_135258449_135258472 | 0.65 | 1.6e-05 | Click! |
Activity profile of Elf3 motif
Sorted Z-values of Elf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_142666816 | 15.26 |
ENSMUST00000105935.1
|
Igf2
|
insulin-like growth factor 2 |
| chr2_-_28084877 | 14.61 |
ENSMUST00000028179.8
ENSMUST00000117486.1 ENSMUST00000135472.1 |
Fcnb
|
ficolin B |
| chr8_+_72761868 | 13.21 |
ENSMUST00000058099.8
|
F2rl3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr14_+_80000292 | 9.53 |
ENSMUST00000088735.3
|
Olfm4
|
olfactomedin 4 |
| chr17_+_29135056 | 8.64 |
ENSMUST00000087942.4
|
Rab44
|
RAB44, member RAS oncogene family |
| chr11_+_87793470 | 7.66 |
ENSMUST00000020779.4
|
Mpo
|
myeloperoxidase |
| chr16_+_17980565 | 7.48 |
ENSMUST00000075371.3
|
Vpreb2
|
pre-B lymphocyte gene 2 |
| chr11_+_87793722 | 7.33 |
ENSMUST00000143021.2
|
Mpo
|
myeloperoxidase |
| chr11_+_115887601 | 7.14 |
ENSMUST00000167507.2
|
Myo15b
|
myosin XVB |
| chr12_+_109544498 | 6.99 |
ENSMUST00000126289.1
|
Meg3
|
maternally expressed 3 |
| chr5_-_43981757 | 6.92 |
ENSMUST00000061299.7
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chrX_-_88760312 | 6.17 |
ENSMUST00000182943.1
|
Gm27000
|
predicted gene, 27000 |
| chr15_-_103255433 | 5.77 |
ENSMUST00000075192.6
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr19_-_17356631 | 5.72 |
ENSMUST00000174236.1
|
Gcnt1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr1_-_128592284 | 5.40 |
ENSMUST00000052172.6
ENSMUST00000142893.1 |
Cxcr4
|
chemokine (C-X-C motif) receptor 4 |
| chr17_+_25471564 | 5.19 |
ENSMUST00000025002.1
|
Tekt4
|
tektin 4 |
| chr17_+_31208049 | 5.04 |
ENSMUST00000173776.1
|
Ubash3a
|
ubiquitin associated and SH3 domain containing, A |
| chr7_+_24777172 | 4.81 |
ENSMUST00000038069.7
|
Ceacam10
|
carcinoembryonic antigen-related cell adhesion molecule 10 |
| chr5_+_115845229 | 4.62 |
ENSMUST00000137952.1
ENSMUST00000148245.1 |
Cit
|
citron |
| chr11_-_97996171 | 4.53 |
ENSMUST00000042971.9
|
Arl5c
|
ADP-ribosylation factor-like 5C |
| chr17_+_35049966 | 4.45 |
ENSMUST00000007257.9
|
Clic1
|
chloride intracellular channel 1 |
| chrX_-_135210672 | 4.25 |
ENSMUST00000033783.1
|
Tceal6
|
transcription elongation factor A (SII)-like 6 |
| chr10_-_6980376 | 4.07 |
ENSMUST00000105617.1
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr11_-_6520894 | 4.05 |
ENSMUST00000003459.3
|
Myo1g
|
myosin IG |
| chr10_+_79927039 | 4.03 |
ENSMUST00000019708.5
ENSMUST00000105377.1 |
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr12_-_65172560 | 4.01 |
ENSMUST00000052201.8
|
Mis18bp1
|
MIS18 binding protein 1 |
| chr1_-_193201435 | 3.92 |
ENSMUST00000043550.4
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr3_+_84666192 | 3.75 |
ENSMUST00000107682.1
|
Tmem154
|
transmembrane protein 154 |
| chr3_+_103832562 | 3.71 |
ENSMUST00000062945.5
|
Bcl2l15
|
BCLl2-like 15 |
| chr7_+_131032061 | 3.69 |
ENSMUST00000084509.3
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr8_+_72189613 | 3.63 |
ENSMUST00000072097.6
|
Hsh2d
|
hematopoietic SH2 domain containing |
| chr1_+_136467958 | 3.53 |
ENSMUST00000047817.6
|
Kif14
|
kinesin family member 14 |
| chr6_+_60944472 | 3.40 |
ENSMUST00000129603.1
|
Mmrn1
|
multimerin 1 |
| chr6_-_129917650 | 3.29 |
ENSMUST00000118060.1
|
Klra5
|
killer cell lectin-like receptor, subfamily A, member 5 |
| chrX_-_8090442 | 3.28 |
ENSMUST00000033505.6
|
Was
|
Wiskott-Aldrich syndrome homolog (human) |
| chr7_-_3677509 | 3.28 |
ENSMUST00000038743.8
|
Tmc4
|
transmembrane channel-like gene family 4 |
| chr2_+_164940742 | 3.17 |
ENSMUST00000137626.1
|
Mmp9
|
matrix metallopeptidase 9 |
| chr10_-_30655859 | 3.11 |
ENSMUST00000092610.4
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr9_-_66919646 | 3.10 |
ENSMUST00000041139.7
|
Rab8b
|
RAB8B, member RAS oncogene family |
| chr1_-_87101590 | 3.08 |
ENSMUST00000113270.2
|
Alpi
|
alkaline phosphatase, intestinal |
| chr1_+_135799833 | 3.08 |
ENSMUST00000148201.1
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chr1_+_170277376 | 3.07 |
ENSMUST00000179976.1
|
Sh2d1b1
|
SH2 domain protein 1B1 |
| chr2_+_131491958 | 3.07 |
ENSMUST00000110181.1
ENSMUST00000110180.1 |
Smox
|
spermine oxidase |
| chr3_-_15332285 | 2.98 |
ENSMUST00000108361.1
|
Gm9733
|
predicted gene 9733 |
| chr3_-_51396528 | 2.95 |
ENSMUST00000038154.5
|
Mgarp
|
mitochondria localized glutamic acid rich protein |
| chr2_+_131491764 | 2.92 |
ENSMUST00000028806.5
ENSMUST00000110179.2 ENSMUST00000110189.2 ENSMUST00000110182.2 ENSMUST00000110183.2 ENSMUST00000110186.2 ENSMUST00000110188.1 |
Smox
|
spermine oxidase |
| chrX_+_48146436 | 2.85 |
ENSMUST00000033427.6
|
Sash3
|
SAM and SH3 domain containing 3 |
| chr2_-_32083783 | 2.72 |
ENSMUST00000056406.6
|
Fam78a
|
family with sequence similarity 78, member A |
| chr17_+_71019503 | 2.68 |
ENSMUST00000024847.7
|
Myom1
|
myomesin 1 |
| chr7_+_30776394 | 2.67 |
ENSMUST00000041703.7
|
Dmkn
|
dermokine |
| chr2_+_101624734 | 2.61 |
ENSMUST00000111227.1
|
Rag2
|
recombination activating gene 2 |
| chr8_-_88636117 | 2.60 |
ENSMUST00000034087.7
|
Snx20
|
sorting nexin 20 |
| chr3_-_51396502 | 2.54 |
ENSMUST00000108046.1
|
Mgarp
|
mitochondria localized glutamic acid rich protein |
| chr3_-_51396716 | 2.51 |
ENSMUST00000141156.1
|
Mgarp
|
mitochondria localized glutamic acid rich protein |
| chr5_-_139736291 | 2.49 |
ENSMUST00000044642.10
|
Micall2
|
MICAL-like 2 |
| chr16_-_19883873 | 2.47 |
ENSMUST00000100083.3
|
A930003A15Rik
|
RIKEN cDNA A930003A15 gene |
| chr1_-_133690100 | 2.47 |
ENSMUST00000169295.1
|
Lax1
|
lymphocyte transmembrane adaptor 1 |
| chr11_-_17008647 | 2.44 |
ENSMUST00000102881.3
|
Plek
|
pleckstrin |
| chr4_-_63403330 | 2.42 |
ENSMUST00000035724.4
|
Akna
|
AT-hook transcription factor |
| chr6_+_29694204 | 2.40 |
ENSMUST00000046750.7
ENSMUST00000115250.3 |
Tspan33
|
tetraspanin 33 |
| chr1_+_166130467 | 2.37 |
ENSMUST00000166860.1
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr8_+_88294204 | 2.37 |
ENSMUST00000098521.2
|
Adcy7
|
adenylate cyclase 7 |
| chr5_-_34637107 | 2.36 |
ENSMUST00000124668.1
ENSMUST00000001109.4 ENSMUST00000155577.1 ENSMUST00000114329.1 |
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chr3_-_89418287 | 2.34 |
ENSMUST00000029679.3
|
Cks1b
|
CDC28 protein kinase 1b |
| chr13_+_44729535 | 2.32 |
ENSMUST00000174068.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr5_-_34637203 | 2.29 |
ENSMUST00000114331.3
|
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chr16_+_10170228 | 2.28 |
ENSMUST00000044103.5
|
Rpl39l
|
ribosomal protein L39-like |
| chr4_+_127172866 | 2.27 |
ENSMUST00000106094.2
|
Dlgap3
|
discs, large (Drosophila) homolog-associated protein 3 |
| chr1_+_166130238 | 2.21 |
ENSMUST00000060833.7
ENSMUST00000166159.1 |
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr4_-_156255327 | 2.20 |
ENSMUST00000179919.1
|
Samd11
|
sterile alpha motif domain containing 11 |
| chr8_-_65129317 | 2.18 |
ENSMUST00000098713.3
|
BC030870
|
cDNA sequence BC030870 |
| chr7_+_30291659 | 2.18 |
ENSMUST00000014065.8
ENSMUST00000150892.1 ENSMUST00000126216.1 |
Clip3
|
CAP-GLY domain containing linker protein 3 |
| chr15_-_66812593 | 2.16 |
ENSMUST00000100572.3
|
Sla
|
src-like adaptor |
| chr16_-_3718105 | 2.10 |
ENSMUST00000023180.7
ENSMUST00000100222.2 |
Mefv
|
Mediterranean fever |
| chr19_-_50678642 | 2.08 |
ENSMUST00000072685.6
ENSMUST00000164039.2 |
Sorcs1
|
VPS10 domain receptor protein SORCS 1 |
| chr15_-_97908261 | 2.08 |
ENSMUST00000023119.8
|
Vdr
|
vitamin D receptor |
| chr1_+_51987139 | 2.08 |
ENSMUST00000168302.1
|
Stat4
|
signal transducer and activator of transcription 4 |
| chr14_+_31208309 | 2.05 |
ENSMUST00000169169.1
|
Tnnc1
|
troponin C, cardiac/slow skeletal |
| chr4_-_11965699 | 2.05 |
ENSMUST00000108301.1
ENSMUST00000095144.3 ENSMUST00000108302.1 |
Pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr5_+_110330697 | 2.01 |
ENSMUST00000112481.1
|
Pole
|
polymerase (DNA directed), epsilon |
| chr6_-_78378851 | 2.00 |
ENSMUST00000089667.1
ENSMUST00000167492.1 |
Reg3d
|
regenerating islet-derived 3 delta |
| chr16_+_19760232 | 1.99 |
ENSMUST00000079780.3
ENSMUST00000164397.1 |
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr13_+_44729794 | 1.98 |
ENSMUST00000172830.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr11_+_11489266 | 1.97 |
ENSMUST00000109678.1
|
4930415F15Rik
|
RIKEN cDNA 4930415F15 gene |
| chr19_+_11404735 | 1.94 |
ENSMUST00000153546.1
|
Ms4a4c
|
membrane-spanning 4-domains, subfamily A, member 4C |
| chr7_-_45238794 | 1.94 |
ENSMUST00000098461.1
ENSMUST00000107797.1 |
Cd37
|
CD37 antigen |
| chr3_-_52104891 | 1.93 |
ENSMUST00000121440.1
|
Maml3
|
mastermind like 3 (Drosophila) |
| chr1_+_40324570 | 1.93 |
ENSMUST00000095020.3
|
Il1rl2
|
interleukin 1 receptor-like 2 |
| chr11_+_32533290 | 1.93 |
ENSMUST00000102821.3
|
Stk10
|
serine/threonine kinase 10 |
| chr12_-_8539545 | 1.92 |
ENSMUST00000095863.3
ENSMUST00000165657.1 |
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chr13_+_13437602 | 1.84 |
ENSMUST00000005532.7
|
Nid1
|
nidogen 1 |
| chr15_+_79516396 | 1.84 |
ENSMUST00000010974.7
|
Kdelr3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr5_-_131538687 | 1.80 |
ENSMUST00000161374.1
|
Auts2
|
autism susceptibility candidate 2 |
| chr10_+_127323700 | 1.79 |
ENSMUST00000069548.5
|
Arhgap9
|
Rho GTPase activating protein 9 |
| chr17_+_32403006 | 1.79 |
ENSMUST00000065921.5
|
A530088E08Rik
|
RIKEN cDNA A530088E08 gene |
| chr11_-_117779605 | 1.78 |
ENSMUST00000143406.1
|
Tmc6
|
transmembrane channel-like gene family 6 |
| chr1_-_156474249 | 1.77 |
ENSMUST00000051396.6
|
Soat1
|
sterol O-acyltransferase 1 |
| chr10_-_117292863 | 1.74 |
ENSMUST00000092162.5
|
Lyz1
|
lysozyme 1 |
| chr7_-_104950441 | 1.73 |
ENSMUST00000179862.1
|
Gm5900
|
predicted pseudogene 5900 |
| chr3_-_105932664 | 1.72 |
ENSMUST00000098758.2
|
I830077J02Rik
|
RIKEN cDNA I830077J02 gene |
| chr7_+_29983948 | 1.72 |
ENSMUST00000148442.1
|
Zfp568
|
zinc finger protein 568 |
| chr9_-_120068263 | 1.72 |
ENSMUST00000064165.3
ENSMUST00000177637.1 |
Cx3cr1
|
chemokine (C-X3-C) receptor 1 |
| chr14_-_20496780 | 1.68 |
ENSMUST00000022353.3
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr1_+_75382114 | 1.67 |
ENSMUST00000113590.1
ENSMUST00000148515.1 |
Speg
|
SPEG complex locus |
| chr3_+_95160449 | 1.66 |
ENSMUST00000090823.1
ENSMUST00000090821.3 |
Sema6c
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr10_+_79988584 | 1.66 |
ENSMUST00000004784.4
ENSMUST00000105374.1 |
Cnn2
|
calponin 2 |
| chrX_+_9350599 | 1.64 |
ENSMUST00000073949.2
|
Gm14501
|
predicted gene 14501 |
| chr4_-_156228540 | 1.64 |
ENSMUST00000105571.2
|
Plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr17_-_47834682 | 1.64 |
ENSMUST00000066368.6
|
Mdfi
|
MyoD family inhibitor |
| chr10_-_23787195 | 1.60 |
ENSMUST00000073926.6
|
Rps12
|
ribosomal protein S12 |
| chr6_-_16898441 | 1.60 |
ENSMUST00000031533.7
|
Tfec
|
transcription factor EC |
| chr1_-_136230289 | 1.59 |
ENSMUST00000150163.1
ENSMUST00000144464.1 |
5730559C18Rik
|
RIKEN cDNA 5730559C18 gene |
| chrX_+_164139321 | 1.56 |
ENSMUST00000112271.3
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr7_+_30763750 | 1.55 |
ENSMUST00000165887.1
ENSMUST00000085691.4 ENSMUST00000085688.4 ENSMUST00000054427.6 |
Dmkn
|
dermokine |
| chr16_+_19760195 | 1.55 |
ENSMUST00000121344.1
|
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr9_-_119209096 | 1.54 |
ENSMUST00000084797.4
|
Slc22a13
|
solute carrier family 22 (organic cation transporter), member 13 |
| chr15_-_9529868 | 1.54 |
ENSMUST00000003981.4
|
Il7r
|
interleukin 7 receptor |
| chr19_-_50678485 | 1.53 |
ENSMUST00000111756.3
|
Sorcs1
|
VPS10 domain receptor protein SORCS 1 |
| chrX_+_11302432 | 1.53 |
ENSMUST00000179428.1
|
Gm14474
|
predicted gene 14474 |
| chr12_-_112860886 | 1.52 |
ENSMUST00000021729.7
|
Gpr132
|
G protein-coupled receptor 132 |
| chr7_+_130865835 | 1.48 |
ENSMUST00000075181.4
ENSMUST00000048180.5 |
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr3_+_88376023 | 1.41 |
ENSMUST00000179946.1
|
Gm21956
|
predicted gene, 21956 |
| chr16_+_92485705 | 1.36 |
ENSMUST00000162181.1
|
Clic6
|
chloride intracellular channel 6 |
| chr6_+_119175247 | 1.35 |
ENSMUST00000112777.2
ENSMUST00000073909.5 |
Dcp1b
|
DCP1 decapping enzyme homolog B (S. cerevisiae) |
| chr3_+_129881149 | 1.34 |
ENSMUST00000061165.7
|
Pla2g12a
|
phospholipase A2, group XIIA |
| chr6_-_145434925 | 1.34 |
ENSMUST00000111708.2
|
Ifltd1
|
intermediate filament tail domain containing 1 |
| chr5_-_123879992 | 1.34 |
ENSMUST00000164267.1
|
Gpr81
|
G protein-coupled receptor 81 |
| chr7_-_125491397 | 1.33 |
ENSMUST00000138616.1
|
Nsmce1
|
non-SMC element 1 homolog (S. cerevisiae) |
| chr8_-_119605199 | 1.33 |
ENSMUST00000093099.6
|
Taf1c
|
TATA box binding protein (Tbp)-associated factor, RNA polymerase I, C |
| chr16_+_92498122 | 1.30 |
ENSMUST00000023670.3
|
Clic6
|
chloride intracellular channel 6 |
| chr3_+_87376381 | 1.28 |
ENSMUST00000163661.1
ENSMUST00000072480.2 ENSMUST00000167200.1 |
Fcrl1
|
Fc receptor-like 1 |
| chr8_+_75214502 | 1.28 |
ENSMUST00000132133.1
|
Rasd2
|
RASD family, member 2 |
| chrX_+_73123068 | 1.26 |
ENSMUST00000179117.1
|
Gm14685
|
predicted gene 14685 |
| chr7_+_142434977 | 1.21 |
ENSMUST00000118276.1
ENSMUST00000105976.1 ENSMUST00000097939.2 |
Syt8
|
synaptotagmin VIII |
| chr3_+_90603767 | 1.20 |
ENSMUST00000001046.5
ENSMUST00000107330.1 |
S100a4
|
S100 calcium binding protein A4 |
| chr5_+_33983437 | 1.20 |
ENSMUST00000114384.1
ENSMUST00000094869.5 ENSMUST00000114383.1 |
Gm1673
|
predicted gene 1673 |
| chr10_-_62231208 | 1.19 |
ENSMUST00000047883.9
|
Tspan15
|
tetraspanin 15 |
| chr6_-_128300738 | 1.19 |
ENSMUST00000143004.1
ENSMUST00000006311.6 ENSMUST00000112157.2 ENSMUST00000133118.1 |
Tead4
|
TEA domain family member 4 |
| chr11_-_76571527 | 1.18 |
ENSMUST00000072740.6
|
Abr
|
active BCR-related gene |
| chr5_+_33983534 | 1.18 |
ENSMUST00000114382.1
|
Gm1673
|
predicted gene 1673 |
| chr8_-_82403203 | 1.18 |
ENSMUST00000034148.6
|
Il15
|
interleukin 15 |
| chrX_+_152781986 | 1.18 |
ENSMUST00000026383.3
|
Gpr143
|
G protein-coupled receptor 143 |
| chr3_-_103791075 | 1.17 |
ENSMUST00000106845.2
ENSMUST00000029438.8 ENSMUST00000121324.1 |
Hipk1
|
homeodomain interacting protein kinase 1 |
| chr1_+_170308802 | 1.13 |
ENSMUST00000056991.5
|
1700015E13Rik
|
RIKEN cDNA 1700015E13 gene |
| chr1_-_161876656 | 1.12 |
ENSMUST00000048377.5
|
Suco
|
SUN domain containing ossification factor |
| chr11_-_103344651 | 1.10 |
ENSMUST00000041385.7
|
Arhgap27
|
Rho GTPase activating protein 27 |
| chr5_+_108065742 | 1.08 |
ENSMUST00000081567.4
ENSMUST00000170319.1 ENSMUST00000112626.1 |
Mtf2
|
metal response element binding transcription factor 2 |
| chr7_+_101361250 | 1.07 |
ENSMUST00000137384.1
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr17_-_17883920 | 1.07 |
ENSMUST00000061516.7
|
Fpr1
|
formyl peptide receptor 1 |
| chr7_-_45759527 | 1.06 |
ENSMUST00000075571.7
|
Sult2b1
|
sulfotransferase family, cytosolic, 2B, member 1 |
| chr2_+_165055668 | 1.06 |
ENSMUST00000081310.4
ENSMUST00000140951.1 |
Cd40
|
CD40 antigen |
| chr7_+_102065713 | 1.05 |
ENSMUST00000094129.2
ENSMUST00000094130.2 ENSMUST00000084843.3 |
Trpc2
|
transient receptor potential cation channel, subfamily C, member 2 |
| chr4_-_83021102 | 1.04 |
ENSMUST00000071708.5
|
Frem1
|
Fras1 related extracellular matrix protein 1 |
| chr7_-_125491586 | 1.04 |
ENSMUST00000033006.7
|
Nsmce1
|
non-SMC element 1 homolog (S. cerevisiae) |
| chr2_-_119229885 | 1.03 |
ENSMUST00000076084.5
|
Ppp1r14d
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr9_-_43116514 | 1.03 |
ENSMUST00000061833.4
|
Tmem136
|
transmembrane protein 136 |
| chr14_+_53709902 | 1.03 |
ENSMUST00000103638.4
|
Trav6-7-dv9
|
T cell receptor alpha variable 6-7-DV9 |
| chr10_+_7667503 | 1.02 |
ENSMUST00000040135.8
|
Nup43
|
nucleoporin 43 |
| chr17_-_25880236 | 1.00 |
ENSMUST00000176696.1
ENSMUST00000095487.5 |
Wfikkn1
|
WAP, FS, Ig, KU, and NTR-containing protein 1 |
| chr7_+_127876796 | 1.00 |
ENSMUST00000131000.1
|
Zfp646
|
zinc finger protein 646 |
| chr11_-_94782500 | 1.00 |
ENSMUST00000162809.2
|
Tmem92
|
transmembrane protein 92 |
| chr11_-_121118677 | 0.97 |
ENSMUST00000039146.3
|
Tex19.2
|
testis expressed gene 19.2 |
| chr14_-_70524068 | 0.97 |
ENSMUST00000022692.3
|
Sftpc
|
surfactant associated protein C |
| chr9_-_32928928 | 0.97 |
ENSMUST00000185169.1
|
RP24-308I2.1
|
RP24-308I2.1 |
| chr1_-_75264195 | 0.94 |
ENSMUST00000027404.5
|
Ptprn
|
protein tyrosine phosphatase, receptor type, N |
| chr14_-_70175397 | 0.89 |
ENSMUST00000143393.1
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr7_-_119793958 | 0.89 |
ENSMUST00000106523.1
ENSMUST00000063902.7 ENSMUST00000150844.1 |
Eri2
|
exoribonuclease 2 |
| chr11_+_77765588 | 0.88 |
ENSMUST00000164315.1
|
Myo18a
|
myosin XVIIIA |
| chrX_+_9283764 | 0.87 |
ENSMUST00000177926.1
|
1700012L04Rik
|
RIKEN cDNA 1700012L04 gene |
| chr9_-_76567092 | 0.87 |
ENSMUST00000183437.1
|
Fam83b
|
family with sequence similarity 83, member B |
| chr16_+_18498768 | 0.86 |
ENSMUST00000167778.1
ENSMUST00000139625.1 ENSMUST00000149035.1 ENSMUST00000090086.4 ENSMUST00000115601.1 ENSMUST00000147739.1 ENSMUST00000146673.1 |
Gnb1l
Gm16314
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like predicted gene 16314 |
| chr17_+_5975740 | 0.86 |
ENSMUST00000115790.1
|
Synj2
|
synaptojanin 2 |
| chrX_-_163761323 | 0.85 |
ENSMUST00000059320.2
|
Rnf138rt1
|
ring finger protein 138, retrogene 1 |
| chr11_+_111066154 | 0.83 |
ENSMUST00000042970.2
|
Kcnj2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr8_+_66386292 | 0.83 |
ENSMUST00000039540.5
ENSMUST00000110253.2 |
March1
|
membrane-associated ring finger (C3HC4) 1 |
| chr4_-_136053343 | 0.82 |
ENSMUST00000102536.4
|
Rpl11
|
ribosomal protein L11 |
| chr5_-_77310049 | 0.81 |
ENSMUST00000047860.8
|
Noa1
|
nitric oxide associated 1 |
| chr7_+_119794102 | 0.81 |
ENSMUST00000084644.2
|
2610020H08Rik
|
RIKEN cDNA 2610020H08 gene |
| chr15_-_36608959 | 0.80 |
ENSMUST00000001809.8
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr17_-_13052280 | 0.78 |
ENSMUST00000091648.2
|
Gpr31b
|
G protein-coupled receptor 31, D17Leh66b region |
| chr4_+_86930691 | 0.77 |
ENSMUST00000164590.1
|
Acer2
|
alkaline ceramidase 2 |
| chr6_+_115134899 | 0.75 |
ENSMUST00000009538.5
ENSMUST00000169345.1 |
Syn2
|
synapsin II |
| chr7_+_119793987 | 0.75 |
ENSMUST00000033218.8
ENSMUST00000106520.1 |
2610020H08Rik
|
RIKEN cDNA 2610020H08 gene |
| chr1_+_170232749 | 0.71 |
ENSMUST00000162752.1
|
Sh2d1b2
|
SH2 domain protein 1B2 |
| chr18_-_37638719 | 0.71 |
ENSMUST00000058635.2
|
Slc25a2
|
solute carrier family 25 (mitochondrial carrier, ornithine transporter) member 2 |
| chr19_-_5912771 | 0.70 |
ENSMUST00000118623.1
|
Dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr7_-_127208423 | 0.70 |
ENSMUST00000120705.1
|
Tbc1d10b
|
TBC1 domain family, member 10b |
| chr13_+_111867931 | 0.70 |
ENSMUST00000128198.1
|
Gm15326
|
predicted gene 15326 |
| chr2_+_165055625 | 0.69 |
ENSMUST00000017799.5
ENSMUST00000073707.2 |
Cd40
|
CD40 antigen |
| chr8_+_104340594 | 0.69 |
ENSMUST00000034343.4
|
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr7_+_102065485 | 0.69 |
ENSMUST00000106950.1
ENSMUST00000146450.1 |
Trpc2
|
transient receptor potential cation channel, subfamily C, member 2 |
| chr7_+_101896817 | 0.68 |
ENSMUST00000143835.1
|
Anapc15
|
anaphase prompoting complex C subunit 15 |
| chr5_+_77310147 | 0.67 |
ENSMUST00000031167.5
|
Polr2b
|
polymerase (RNA) II (DNA directed) polypeptide B |
| chr5_+_17574268 | 0.66 |
ENSMUST00000030568.7
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr10_-_117238665 | 0.66 |
ENSMUST00000020392.4
|
9530003J23Rik
|
RIKEN cDNA 9530003J23 gene |
| chr12_-_80260356 | 0.64 |
ENSMUST00000021554.8
|
Actn1
|
actinin, alpha 1 |
| chr3_+_98222148 | 0.64 |
ENSMUST00000029469.4
|
Reg4
|
regenerating islet-derived family, member 4 |
| chr12_+_83950608 | 0.64 |
ENSMUST00000053744.7
|
2410016O06Rik
|
RIKEN cDNA 2410016O06 gene |
| chr11_+_98383811 | 0.63 |
ENSMUST00000008021.2
|
Tcap
|
titin-cap |
| chr12_+_65075582 | 0.63 |
ENSMUST00000058889.4
|
Fancm
|
Fanconi anemia, complementation group M |
Network of associatons between targets according to the STRING database.
First level regulatory network of Elf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 15.0 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 3.7 | 14.6 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 1.9 | 15.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 1.3 | 5.4 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 1.2 | 3.5 | GO:0033624 | negative regulation of integrin activation(GO:0033624) |
| 1.1 | 3.2 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 1.1 | 4.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 1.0 | 5.2 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 1.0 | 8.0 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 1.0 | 15.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.9 | 4.7 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.9 | 2.6 | GO:0002358 | B cell homeostatic proliferation(GO:0002358) |
| 0.9 | 6.9 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.9 | 6.0 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.8 | 4.0 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.8 | 3.1 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.7 | 3.6 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.7 | 2.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.7 | 2.0 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.6 | 1.8 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.6 | 1.2 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.5 | 3.3 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.5 | 2.1 | GO:0071608 | macrophage inflammatory protein-1 alpha production(GO:0071608) |
| 0.5 | 2.1 | GO:0038183 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) bile acid signaling pathway(GO:0038183) |
| 0.5 | 5.8 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.4 | 5.7 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.4 | 1.6 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) positive regulation of gap junction assembly(GO:1903598) |
| 0.4 | 3.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.3 | 1.7 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.3 | 1.7 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.3 | 2.2 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.3 | 4.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.3 | 0.8 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.3 | 0.8 | GO:0048866 | endodermal cell fate determination(GO:0007493) stem cell fate specification(GO:0048866) regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) regulation of cardiac cell fate specification(GO:2000043) |
| 0.3 | 1.8 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.3 | 3.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.3 | 0.8 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.2 | 1.2 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.2 | 0.7 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.2 | 1.2 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.2 | 7.0 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.2 | 0.9 | GO:1903028 | asymmetric Golgi ribbon formation(GO:0090164) regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.2 | 5.0 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.2 | 1.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 | 1.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.2 | 3.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 4.6 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.2 | 1.0 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.2 | 3.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 1.9 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.2 | 3.1 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.2 | 1.5 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.2 | 1.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 | 1.8 | GO:0042983 | very-low-density lipoprotein particle assembly(GO:0034379) amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.2 | 1.9 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 1.3 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 1.2 | GO:1903056 | regulation of melanosome organization(GO:1903056) |
| 0.2 | 2.0 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.2 | 3.3 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.2 | 0.8 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.2 | 0.8 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.2 | 0.8 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 | 2.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 2.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 1.8 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 2.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 2.0 | GO:0035872 | nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway(GO:0035872) |
| 0.1 | 8.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.1 | GO:0032805 | positive regulation of low-density lipoprotein particle receptor catabolic process(GO:0032805) |
| 0.1 | 0.5 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.1 | 0.6 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 0.4 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.6 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 7.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.3 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.1 | 1.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.6 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.6 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.3 | GO:0036363 | transforming growth factor beta activation(GO:0036363) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.7 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.7 | GO:0003350 | pulmonary myocardium development(GO:0003350) dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 2.4 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 0.3 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 0.3 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 1.2 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 1.2 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 2.0 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 0.5 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 1.7 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.1 | 7.5 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.1 | 0.2 | GO:0040010 | positive regulation of growth rate(GO:0040010) |
| 0.1 | 0.4 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 1.6 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 0.5 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.1 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.2 | GO:0046370 | fructose biosynthetic process(GO:0046370) |
| 0.1 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.6 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 2.4 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.6 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 0.5 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 2.5 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 1.5 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 2.2 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.9 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 1.9 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.4 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 1.1 | GO:0044253 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 | 0.8 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 3.5 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 1.5 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.2 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.0 | 1.0 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 1.1 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.4 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.4 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 0.9 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.8 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.3 | GO:2001269 | negative regulation by host of viral transcription(GO:0043922) positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 1.7 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 1.0 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.4 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.0 | 0.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.6 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.0 | 1.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 14.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 1.0 | 9.5 | GO:0042581 | specific granule(GO:0042581) |
| 0.9 | 15.0 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.4 | 2.0 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.4 | 2.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.3 | 2.1 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.3 | 2.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.3 | 2.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.3 | 3.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 1.7 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.3 | 4.0 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.3 | 1.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.3 | 8.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.3 | 3.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 0.6 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.2 | 3.1 | GO:0051286 | cell tip(GO:0051286) |
| 0.2 | 5.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 1.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 5.2 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 4.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.8 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 2.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 8.3 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 7.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 3.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 5.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 2.7 | GO:0032982 | muscle myosin complex(GO:0005859) myosin filament(GO:0032982) |
| 0.1 | 1.0 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 2.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.0 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 1.8 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 1.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.9 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 5.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.3 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 1.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.7 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.4 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 8.1 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 2.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 6.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 5.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.8 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.6 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 2.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 5.9 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 1.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 1.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 2.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 2.6 | GO:0012506 | vesicle membrane(GO:0012506) |
| 0.0 | 0.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.8 | GO:0005875 | microtubule associated complex(GO:0005875) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 13.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 2.0 | 6.0 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 1.9 | 5.7 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 1.6 | 14.6 | GO:0033691 | sialic acid binding(GO:0033691) |
| 1.2 | 3.5 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.9 | 4.7 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.9 | 2.7 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.8 | 5.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.7 | 2.1 | GO:1902271 | lithocholic acid binding(GO:1902121) D3 vitamins binding(GO:1902271) |
| 0.6 | 1.9 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.6 | 2.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.5 | 2.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.5 | 1.8 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.4 | 15.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.4 | 3.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.4 | 3.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.4 | 2.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.4 | 2.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.3 | 1.7 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.3 | 10.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.3 | 2.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.3 | 2.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.3 | 0.8 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.3 | 2.0 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.2 | 15.0 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.2 | 1.6 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.2 | 0.9 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.2 | 6.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.2 | 10.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 0.4 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.2 | 1.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 1.1 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.1 | 2.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 2.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 1.2 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.1 | 0.7 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 2.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.1 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.1 | 0.6 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 5.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 2.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 3.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 3.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 3.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.6 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.1 | 0.8 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 1.4 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 4.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 0.8 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.1 | 0.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.8 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 3.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 3.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 6.0 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 1.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.7 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 2.5 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 1.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.9 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.1 | 0.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 1.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.2 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.1 | 0.1 | GO:0034617 | nitric-oxide synthase activity(GO:0004517) tetrahydrobiopterin binding(GO:0034617) |
| 0.1 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 2.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.6 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 3.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 1.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.0 | 1.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.6 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 1.0 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 1.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.9 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.8 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 2.4 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 7.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0052851 | ferric-chelate reductase activity(GO:0000293) cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.7 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 2.4 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 5.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 11.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 17.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 9.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 15.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 4.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.8 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 2.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 4.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 2.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 3.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 2.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 3.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 6.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 2.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.5 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.3 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.7 | 15.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 6.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 11.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.2 | 2.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 2.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.2 | 2.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 8.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 2.1 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 3.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.9 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 5.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 4.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 2.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 3.3 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 1.0 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 2.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 4.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 1.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 3.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |