Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
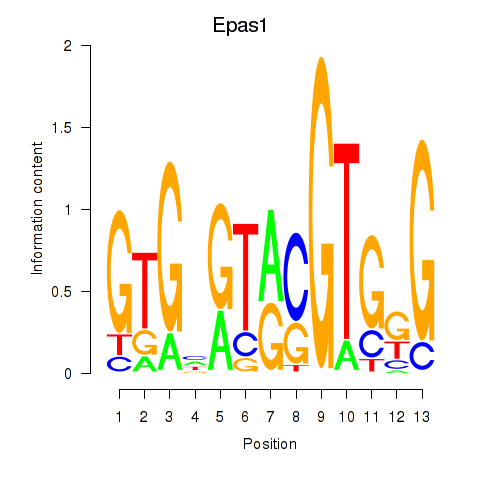
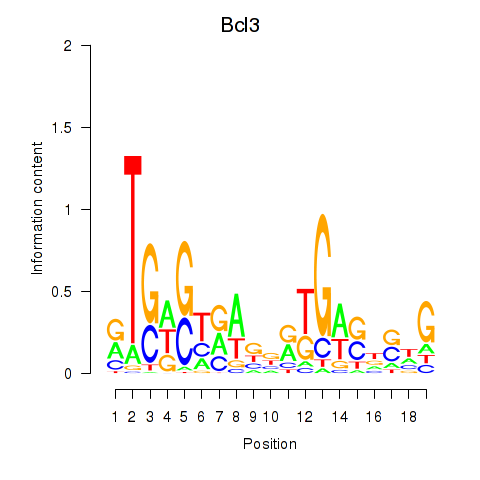
Results for Epas1_Bcl3
Z-value: 1.32
Transcription factors associated with Epas1_Bcl3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Epas1
|
ENSMUSG00000024140.9 | endothelial PAS domain protein 1 |
|
Bcl3
|
ENSMUSG00000053175.10 | B cell leukemia/lymphoma 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Epas1 | mm10_v2_chr17_+_86753900_86753914 | 0.33 | 4.9e-02 | Click! |
| Bcl3 | mm10_v2_chr7_-_19822698_19822776 | -0.09 | 6.0e-01 | Click! |
Activity profile of Epas1_Bcl3 motif
Sorted Z-values of Epas1_Bcl3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_28560704 | 13.10 |
ENSMUST00000114785.1
ENSMUST00000025062.3 |
Clps
|
colipase, pancreatic |
| chr14_-_60177482 | 12.25 |
ENSMUST00000140924.1
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr14_+_55765956 | 9.80 |
ENSMUST00000057569.3
|
Ltb4r1
|
leukotriene B4 receptor 1 |
| chr8_+_79028587 | 8.84 |
ENSMUST00000119254.1
|
Zfp827
|
zinc finger protein 827 |
| chr5_-_134747241 | 8.59 |
ENSMUST00000015138.9
|
Eln
|
elastin |
| chr10_+_127725392 | 7.94 |
ENSMUST00000026466.3
|
Tac2
|
tachykinin 2 |
| chr3_+_51661167 | 7.73 |
ENSMUST00000099106.3
|
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr8_+_79028317 | 6.21 |
ENSMUST00000087927.4
ENSMUST00000098614.2 |
Zfp827
|
zinc finger protein 827 |
| chr3_-_132950043 | 5.77 |
ENSMUST00000117164.1
ENSMUST00000093971.4 ENSMUST00000042729.9 ENSMUST00000042744.9 ENSMUST00000117811.1 |
Npnt
|
nephronectin |
| chr3_+_51661209 | 4.89 |
ENSMUST00000161590.1
|
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr19_+_47228804 | 4.38 |
ENSMUST00000111807.3
|
Neurl1a
|
neuralized homolog 1A (Drosophila) |
| chr4_-_134018829 | 4.35 |
ENSMUST00000051674.2
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr6_+_145145473 | 4.24 |
ENSMUST00000156849.1
ENSMUST00000132948.1 |
Lrmp
|
lymphoid-restricted membrane protein |
| chr1_+_75507077 | 4.21 |
ENSMUST00000037330.4
|
Inha
|
inhibin alpha |
| chr11_+_95010277 | 4.07 |
ENSMUST00000124735.1
|
Samd14
|
sterile alpha motif domain containing 14 |
| chr7_-_100863373 | 3.78 |
ENSMUST00000142885.1
ENSMUST00000008462.3 |
Relt
|
RELT tumor necrosis factor receptor |
| chr12_+_102554966 | 3.65 |
ENSMUST00000021610.5
|
Chga
|
chromogranin A |
| chr11_-_102365111 | 3.63 |
ENSMUST00000006749.9
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr6_+_36388055 | 3.40 |
ENSMUST00000172278.1
|
Chrm2
|
cholinergic receptor, muscarinic 2, cardiac |
| chr4_+_17853451 | 3.31 |
ENSMUST00000029881.3
|
Mmp16
|
matrix metallopeptidase 16 |
| chr4_+_49059256 | 3.06 |
ENSMUST00000076670.2
|
E130309F12Rik
|
RIKEN cDNA E130309F12 gene |
| chr13_+_55369732 | 3.02 |
ENSMUST00000063771.7
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr14_-_54994541 | 3.00 |
ENSMUST00000153783.1
ENSMUST00000168485.1 ENSMUST00000102803.3 |
Myh7
|
myosin, heavy polypeptide 7, cardiac muscle, beta |
| chr6_+_128399766 | 2.99 |
ENSMUST00000001561.5
|
Nrip2
|
nuclear receptor interacting protein 2 |
| chr17_+_36869567 | 2.93 |
ENSMUST00000060524.9
|
Trim10
|
tripartite motif-containing 10 |
| chr14_-_70630149 | 2.84 |
ENSMUST00000022694.9
|
Dmtn
|
dematin actin binding protein |
| chr12_-_16800674 | 2.78 |
ENSMUST00000162112.1
|
Greb1
|
gene regulated by estrogen in breast cancer protein |
| chr10_+_79879614 | 2.77 |
ENSMUST00000006679.8
|
Prtn3
|
proteinase 3 |
| chr9_+_65101453 | 2.74 |
ENSMUST00000077696.6
ENSMUST00000035499.4 ENSMUST00000166273.1 |
Igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr15_+_80173642 | 2.63 |
ENSMUST00000044970.6
|
Mgat3
|
mannoside acetylglucosaminyltransferase 3 |
| chr15_+_81936911 | 2.54 |
ENSMUST00000135663.1
|
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr2_+_105127200 | 2.51 |
ENSMUST00000139585.1
|
Wt1
|
Wilms tumor 1 homolog |
| chr10_+_42860348 | 2.50 |
ENSMUST00000063063.7
|
Scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr11_-_101785252 | 2.41 |
ENSMUST00000164750.1
ENSMUST00000107176.1 ENSMUST00000017868.6 |
Etv4
|
ets variant gene 4 (E1A enhancer binding protein, E1AF) |
| chr8_-_48555846 | 2.41 |
ENSMUST00000110345.1
ENSMUST00000110343.1 |
Tenm3
|
teneurin transmembrane protein 3 |
| chr9_+_62858085 | 2.40 |
ENSMUST00000034777.6
ENSMUST00000163820.1 |
Calml4
|
calmodulin-like 4 |
| chr7_+_122671378 | 2.35 |
ENSMUST00000182563.1
|
Cacng3
|
calcium channel, voltage-dependent, gamma subunit 3 |
| chr3_-_98339921 | 2.27 |
ENSMUST00000065793.5
|
Phgdh
|
3-phosphoglycerate dehydrogenase |
| chr10_+_17796256 | 2.24 |
ENSMUST00000037964.6
|
Txlnb
|
taxilin beta |
| chr7_+_79660196 | 2.19 |
ENSMUST00000035977.7
|
Ticrr
|
TOPBP1-interacting checkpoint and replication regulator |
| chr11_+_95009852 | 2.17 |
ENSMUST00000055947.3
|
Samd14
|
sterile alpha motif domain containing 14 |
| chr7_-_126369543 | 2.15 |
ENSMUST00000032997.6
|
Lat
|
linker for activation of T cells |
| chr17_-_35066170 | 2.15 |
ENSMUST00000174190.1
ENSMUST00000097337.1 |
AU023871
|
expressed sequence AU023871 |
| chr7_+_99535652 | 2.14 |
ENSMUST00000032995.8
ENSMUST00000162404.1 |
Arrb1
|
arrestin, beta 1 |
| chr3_-_94473591 | 2.13 |
ENSMUST00000029785.3
|
Riiad1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 |
| chr2_+_4300462 | 2.12 |
ENSMUST00000175669.1
|
Frmd4a
|
FERM domain containing 4A |
| chr3_-_127225847 | 2.09 |
ENSMUST00000182726.1
ENSMUST00000182779.1 |
Ank2
|
ankyrin 2, brain |
| chr6_+_128399881 | 2.06 |
ENSMUST00000120405.1
|
Nrip2
|
nuclear receptor interacting protein 2 |
| chr4_+_130913264 | 2.05 |
ENSMUST00000156225.1
ENSMUST00000156742.1 |
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr2_+_118814195 | 2.03 |
ENSMUST00000110842.1
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr10_+_42860776 | 2.03 |
ENSMUST00000105494.1
|
Scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr6_-_128826305 | 1.99 |
ENSMUST00000174544.1
ENSMUST00000172887.1 ENSMUST00000032472.4 |
Klrb1b
|
killer cell lectin-like receptor subfamily B member 1B |
| chr9_+_110763646 | 1.99 |
ENSMUST00000079784.7
|
Myl3
|
myosin, light polypeptide 3 |
| chr12_-_14152038 | 1.95 |
ENSMUST00000020926.6
|
Fam84a
|
family with sequence similarity 84, member A |
| chr2_+_118814237 | 1.94 |
ENSMUST00000028803.7
ENSMUST00000126045.1 |
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr11_+_69095217 | 1.91 |
ENSMUST00000101004.2
|
Per1
|
period circadian clock 1 |
| chr9_+_78615501 | 1.89 |
ENSMUST00000093812.4
|
Cd109
|
CD109 antigen |
| chr2_+_118813995 | 1.87 |
ENSMUST00000134661.1
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr1_-_155232710 | 1.86 |
ENSMUST00000035914.3
|
BC034090
|
cDNA sequence BC034090 |
| chr7_+_110772604 | 1.81 |
ENSMUST00000005829.6
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr11_-_97996171 | 1.72 |
ENSMUST00000042971.9
|
Arl5c
|
ADP-ribosylation factor-like 5C |
| chr4_-_126325672 | 1.71 |
ENSMUST00000102616.1
|
Tekt2
|
tektin 2 |
| chr9_-_40346290 | 1.71 |
ENSMUST00000121357.1
|
Gramd1b
|
GRAM domain containing 1B |
| chr5_-_137072254 | 1.70 |
ENSMUST00000077523.3
ENSMUST00000041388.4 |
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1 |
| chr14_-_70524068 | 1.66 |
ENSMUST00000022692.3
|
Sftpc
|
surfactant associated protein C |
| chr1_-_171607321 | 1.66 |
ENSMUST00000111277.1
ENSMUST00000004827.7 |
Ly9
|
lymphocyte antigen 9 |
| chr17_-_29237759 | 1.63 |
ENSMUST00000137727.1
ENSMUST00000024805.7 |
Cpne5
|
copine V |
| chr8_+_70152754 | 1.62 |
ENSMUST00000072500.6
ENSMUST00000164040.1 ENSMUST00000110146.2 ENSMUST00000110143.1 ENSMUST00000110141.2 ENSMUST00000110140.1 |
2310045N01Rik
Mef2b
|
RIKEN cDNA 2310045N01 gene myocyte enhancer factor 2B |
| chr11_-_70352029 | 1.62 |
ENSMUST00000019068.6
|
Alox15
|
arachidonate 15-lipoxygenase |
| chr7_+_122671401 | 1.61 |
ENSMUST00000182095.1
|
Cacng3
|
calcium channel, voltage-dependent, gamma subunit 3 |
| chr5_+_27261916 | 1.60 |
ENSMUST00000101471.3
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr7_+_110768169 | 1.57 |
ENSMUST00000170374.1
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr5_-_67847400 | 1.56 |
ENSMUST00000113652.1
ENSMUST00000113651.1 ENSMUST00000037380.8 |
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
| chr9_+_36832684 | 1.55 |
ENSMUST00000034630.8
|
Fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr17_+_56304313 | 1.54 |
ENSMUST00000113035.1
ENSMUST00000113039.2 ENSMUST00000142387.1 |
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr18_+_49979427 | 1.52 |
ENSMUST00000148989.2
|
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr14_-_76556662 | 1.51 |
ENSMUST00000064517.7
|
Serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr3_-_104818266 | 1.51 |
ENSMUST00000168015.1
|
Mov10
|
Moloney leukemia virus 10 |
| chr9_-_102354685 | 1.50 |
ENSMUST00000035129.7
ENSMUST00000085169.5 |
Ephb1
|
Eph receptor B1 |
| chr17_-_78906899 | 1.50 |
ENSMUST00000042683.6
ENSMUST00000169544.1 |
Sult6b1
|
sulfotransferase family, cytosolic, 6B, member 1 |
| chr2_+_174760619 | 1.47 |
ENSMUST00000029030.2
|
Edn3
|
endothelin 3 |
| chr8_+_123332676 | 1.46 |
ENSMUST00000010298.6
|
Spire2
|
spire homolog 2 (Drosophila) |
| chr10_+_42860648 | 1.45 |
ENSMUST00000105495.1
|
Scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr2_+_135659625 | 1.43 |
ENSMUST00000134310.1
|
Plcb4
|
phospholipase C, beta 4 |
| chr4_+_130913120 | 1.38 |
ENSMUST00000151698.1
|
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr19_+_16956110 | 1.37 |
ENSMUST00000087689.4
|
Prune2
|
prune homolog 2 (Drosophila) |
| chr14_-_31577318 | 1.33 |
ENSMUST00000112027.2
|
Colq
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr7_-_141117772 | 1.31 |
ENSMUST00000067836.7
|
Ano9
|
anoctamin 9 |
| chr3_-_152668135 | 1.30 |
ENSMUST00000045262.6
|
Ak5
|
adenylate kinase 5 |
| chr11_-_70255329 | 1.30 |
ENSMUST00000108574.2
ENSMUST00000000329.2 |
Alox12
|
arachidonate 12-lipoxygenase |
| chr3_+_105452326 | 1.29 |
ENSMUST00000098761.3
|
Kcnd3
|
potassium voltage-gated channel, Shal-related family, member 3 |
| chr17_-_12868126 | 1.28 |
ENSMUST00000089015.3
|
Mas1
|
MAS1 oncogene |
| chr9_-_44234014 | 1.27 |
ENSMUST00000037644.6
|
Cbl
|
Casitas B-lineage lymphoma |
| chr5_-_30461902 | 1.27 |
ENSMUST00000133509.1
ENSMUST00000074171.6 ENSMUST00000114747.2 ENSMUST00000144125.1 |
Otof
|
otoferlin |
| chr3_-_104818539 | 1.27 |
ENSMUST00000106774.1
ENSMUST00000106775.1 ENSMUST00000166979.1 ENSMUST00000136148.1 |
Mov10
|
Moloney leukemia virus 10 |
| chr14_+_116925516 | 1.26 |
ENSMUST00000125435.1
|
Gpc6
|
glypican 6 |
| chr14_+_116925379 | 1.25 |
ENSMUST00000088483.3
|
Gpc6
|
glypican 6 |
| chr14_+_84443553 | 1.25 |
ENSMUST00000071370.5
|
Pcdh17
|
protocadherin 17 |
| chr4_-_126325641 | 1.25 |
ENSMUST00000131113.1
|
Tekt2
|
tektin 2 |
| chr3_-_123034943 | 1.23 |
ENSMUST00000029761.7
|
Myoz2
|
myozenin 2 |
| chr6_-_29179584 | 1.22 |
ENSMUST00000159200.1
|
Prrt4
|
proline-rich transmembrane protein 4 |
| chr1_-_171607378 | 1.22 |
ENSMUST00000068878.7
|
Ly9
|
lymphocyte antigen 9 |
| chr4_+_47091909 | 1.20 |
ENSMUST00000045041.5
ENSMUST00000107744.1 |
Galnt12
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 |
| chr2_+_143546144 | 1.20 |
ENSMUST00000028905.9
|
Pcsk2
|
proprotein convertase subtilisin/kexin type 2 |
| chr18_+_4634912 | 1.17 |
ENSMUST00000037029.5
|
9430020K01Rik
|
RIKEN cDNA 9430020K01 gene |
| chr13_-_28953690 | 1.17 |
ENSMUST00000067230.5
|
Sox4
|
SRY-box containing gene 4 |
| chr2_+_106695594 | 1.17 |
ENSMUST00000016530.7
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr2_+_27886416 | 1.16 |
ENSMUST00000028280.7
|
Col5a1
|
collagen, type V, alpha 1 |
| chr9_+_21368014 | 1.16 |
ENSMUST00000067646.4
ENSMUST00000115414.1 |
Ilf3
|
interleukin enhancer binding factor 3 |
| chr18_+_49979514 | 1.15 |
ENSMUST00000179937.1
|
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr1_-_192855723 | 1.13 |
ENSMUST00000155579.1
|
Sertad4
|
SERTA domain containing 4 |
| chr10_-_12964252 | 1.13 |
ENSMUST00000163425.1
ENSMUST00000042861.5 |
Stx11
|
syntaxin 11 |
| chr18_+_21072329 | 1.11 |
ENSMUST00000082235.4
|
Mep1b
|
meprin 1 beta |
| chr7_+_99535439 | 1.11 |
ENSMUST00000098266.2
ENSMUST00000179755.1 |
Arrb1
|
arrestin, beta 1 |
| chr2_+_105126505 | 1.10 |
ENSMUST00000143043.1
|
Wt1
|
Wilms tumor 1 homolog |
| chr8_-_116732991 | 1.09 |
ENSMUST00000109102.2
|
Cdyl2
|
chromodomain protein, Y chromosome-like 2 |
| chr17_+_29093763 | 1.06 |
ENSMUST00000023829.6
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr8_+_68880491 | 1.06 |
ENSMUST00000015712.8
|
Lpl
|
lipoprotein lipase |
| chr3_-_127225917 | 1.05 |
ENSMUST00000182064.1
ENSMUST00000182662.1 |
Ank2
|
ankyrin 2, brain |
| chr15_-_56694525 | 1.04 |
ENSMUST00000050544.7
|
Has2
|
hyaluronan synthase 2 |
| chr15_+_82275197 | 1.03 |
ENSMUST00000116423.1
|
Sept3
|
septin 3 |
| chr15_+_81936753 | 1.03 |
ENSMUST00000038757.7
|
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr17_+_34608997 | 1.02 |
ENSMUST00000114140.3
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| chr3_+_134236483 | 1.02 |
ENSMUST00000181904.1
ENSMUST00000053048.9 |
Cxxc4
|
CXXC finger 4 |
| chr8_+_82863351 | 1.00 |
ENSMUST00000078525.5
|
Rnf150
|
ring finger protein 150 |
| chr15_+_99670574 | 1.00 |
ENSMUST00000023758.7
|
Asic1
|
acid-sensing (proton-gated) ion channel 1 |
| chrX_-_102250940 | 0.99 |
ENSMUST00000134887.1
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chrX_+_69360294 | 0.98 |
ENSMUST00000033532.6
|
Aff2
|
AF4/FMR2 family, member 2 |
| chr19_+_23758819 | 0.96 |
ENSMUST00000025830.7
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr11_+_3983636 | 0.95 |
ENSMUST00000078757.1
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr3_-_116253467 | 0.94 |
ENSMUST00000090473.5
|
Gpr88
|
G-protein coupled receptor 88 |
| chr19_-_41743665 | 0.94 |
ENSMUST00000025993.3
|
Slit1
|
slit homolog 1 (Drosophila) |
| chrX_+_144688907 | 0.94 |
ENSMUST00000112843.1
|
Zcchc16
|
zinc finger, CCHC domain containing 16 |
| chr5_+_81021202 | 0.93 |
ENSMUST00000117253.1
ENSMUST00000120128.1 |
Lphn3
|
latrophilin 3 |
| chr4_-_88033328 | 0.93 |
ENSMUST00000078090.5
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr2_+_153345809 | 0.93 |
ENSMUST00000109790.1
|
Asxl1
|
additional sex combs like 1 |
| chr8_+_83997613 | 0.90 |
ENSMUST00000095228.3
|
Samd1
|
sterile alpha motif domain containing 1 |
| chr19_+_6400523 | 0.89 |
ENSMUST00000146831.1
ENSMUST00000035716.8 ENSMUST00000138555.1 ENSMUST00000167240.1 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr14_+_116925291 | 0.88 |
ENSMUST00000078849.4
|
Gpc6
|
glypican 6 |
| chr11_+_34314757 | 0.88 |
ENSMUST00000165963.1
ENSMUST00000093192.3 |
Fam196b
|
family with sequence similarity 196, member B |
| chr16_-_44558879 | 0.88 |
ENSMUST00000114634.1
|
Boc
|
biregional cell adhesion molecule-related/down-regulated by oncogenes (Cdon) binding protein |
| chr19_-_31765027 | 0.87 |
ENSMUST00000065067.6
|
Prkg1
|
protein kinase, cGMP-dependent, type I |
| chr5_+_108132885 | 0.85 |
ENSMUST00000047677.7
|
Ccdc18
|
coiled-coil domain containing 18 |
| chr11_+_105292637 | 0.84 |
ENSMUST00000100335.3
ENSMUST00000021038.4 |
Mrc2
|
mannose receptor, C type 2 |
| chr6_+_119175247 | 0.84 |
ENSMUST00000112777.2
ENSMUST00000073909.5 |
Dcp1b
|
DCP1 decapping enzyme homolog B (S. cerevisiae) |
| chr5_+_26904682 | 0.83 |
ENSMUST00000120555.1
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr6_+_125071277 | 0.83 |
ENSMUST00000140346.2
ENSMUST00000171989.1 |
Lpar5
|
lysophosphatidic acid receptor 5 |
| chr9_+_107554633 | 0.82 |
ENSMUST00000010211.4
|
Rassf1
|
Ras association (RalGDS/AF-6) domain family member 1 |
| chr11_+_71750680 | 0.81 |
ENSMUST00000021168.7
|
Wscd1
|
WSC domain containing 1 |
| chr5_+_81021583 | 0.79 |
ENSMUST00000121707.1
|
Lphn3
|
latrophilin 3 |
| chr17_+_35135463 | 0.78 |
ENSMUST00000173535.1
ENSMUST00000173952.1 |
Bag6
|
BCL2-associated athanogene 6 |
| chr17_+_24038144 | 0.78 |
ENSMUST00000059482.5
|
Prss27
|
protease, serine, 27 |
| chr17_+_48932368 | 0.78 |
ENSMUST00000046254.2
|
Lrfn2
|
leucine rich repeat and fibronectin type III domain containing 2 |
| chr17_+_8525369 | 0.78 |
ENSMUST00000115715.1
|
Pde10a
|
phosphodiesterase 10A |
| chr18_+_22345089 | 0.77 |
ENSMUST00000120223.1
ENSMUST00000097655.3 |
Asxl3
|
additional sex combs like 3 (Drosophila) |
| chr19_-_46044914 | 0.76 |
ENSMUST00000026252.7
|
Ldb1
|
LIM domain binding 1 |
| chr1_+_87470258 | 0.76 |
ENSMUST00000027476.4
|
3110079O15Rik
|
RIKEN cDNA 3110079O15 gene |
| chr8_+_110721462 | 0.76 |
ENSMUST00000052457.8
|
Mtss1l
|
metastasis suppressor 1-like |
| chr12_-_3357012 | 0.76 |
ENSMUST00000180719.1
|
Gm26520
|
predicted gene, 26520 |
| chr19_-_50678642 | 0.75 |
ENSMUST00000072685.6
ENSMUST00000164039.2 |
Sorcs1
|
VPS10 domain receptor protein SORCS 1 |
| chr3_-_105932664 | 0.75 |
ENSMUST00000098758.2
|
I830077J02Rik
|
RIKEN cDNA I830077J02 gene |
| chr3_-_30509752 | 0.74 |
ENSMUST00000172697.1
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr5_+_86071734 | 0.74 |
ENSMUST00000031171.7
|
Stap1
|
signal transducing adaptor family member 1 |
| chr3_+_28263563 | 0.73 |
ENSMUST00000160307.2
ENSMUST00000159680.2 ENSMUST00000160518.1 ENSMUST00000162485.1 ENSMUST00000159308.1 ENSMUST00000162777.1 ENSMUST00000161964.1 |
Tnik
|
TRAF2 and NCK interacting kinase |
| chr3_+_105904377 | 0.73 |
ENSMUST00000000574.1
|
Adora3
|
adenosine A3 receptor |
| chr11_-_76468396 | 0.73 |
ENSMUST00000065028.7
|
Abr
|
active BCR-related gene |
| chr17_+_78508063 | 0.72 |
ENSMUST00000024880.9
|
Vit
|
vitrin |
| chr5_-_137613759 | 0.72 |
ENSMUST00000155251.1
ENSMUST00000124693.1 |
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr19_-_4615453 | 0.71 |
ENSMUST00000053597.2
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr10_+_128225830 | 0.70 |
ENSMUST00000026455.7
|
Mip
|
major intrinsic protein of eye lens fiber |
| chr8_-_104631312 | 0.70 |
ENSMUST00000034351.6
|
Rrad
|
Ras-related associated with diabetes |
| chr3_+_103074009 | 0.69 |
ENSMUST00000090715.6
|
Ampd1
|
adenosine monophosphate deaminase 1 |
| chr19_+_27217011 | 0.68 |
ENSMUST00000164746.1
ENSMUST00000172302.1 |
Vldlr
|
very low density lipoprotein receptor |
| chr7_+_31059342 | 0.68 |
ENSMUST00000039775.7
|
Lgi4
|
leucine-rich repeat LGI family, member 4 |
| chr1_-_171059390 | 0.68 |
ENSMUST00000164044.1
ENSMUST00000169017.1 |
Fcgr3
|
Fc receptor, IgG, low affinity III |
| chr18_+_65581704 | 0.68 |
ENSMUST00000182979.1
|
Zfp532
|
zinc finger protein 532 |
| chr9_-_44251464 | 0.67 |
ENSMUST00000034618.4
|
Pdzd3
|
PDZ domain containing 3 |
| chr9_+_120092106 | 0.66 |
ENSMUST00000048777.2
|
Ccr8
|
chemokine (C-C motif) receptor 8 |
| chr13_+_91461050 | 0.66 |
ENSMUST00000004094.8
ENSMUST00000042122.8 |
Ssbp2
|
single-stranded DNA binding protein 2 |
| chr2_+_156421048 | 0.66 |
ENSMUST00000109574.1
|
Epb4.1l1
|
erythrocyte protein band 4.1-like 1 |
| chr10_+_80264942 | 0.65 |
ENSMUST00000105362.1
ENSMUST00000105361.3 |
Dazap1
|
DAZ associated protein 1 |
| chr2_+_156421083 | 0.65 |
ENSMUST00000125153.2
ENSMUST00000103136.1 ENSMUST00000109577.2 |
Epb4.1l1
|
erythrocyte protein band 4.1-like 1 |
| chr19_-_53464721 | 0.64 |
ENSMUST00000180489.1
|
5830416P10Rik
|
RIKEN cDNA 5830416P10 gene |
| chr9_+_112227443 | 0.63 |
ENSMUST00000161216.1
|
2310075C17Rik
|
RIKEN cDNA 2310075C17 gene |
| chr18_-_32559914 | 0.63 |
ENSMUST00000174000.1
ENSMUST00000174459.1 |
Gypc
|
glycophorin C |
| chr4_+_48045144 | 0.63 |
ENSMUST00000030025.3
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr12_-_86079019 | 0.63 |
ENSMUST00000003687.6
|
Tgfb3
|
transforming growth factor, beta 3 |
| chr2_+_147364989 | 0.62 |
ENSMUST00000109968.2
|
Pax1
|
paired box gene 1 |
| chr5_-_67847360 | 0.62 |
ENSMUST00000072971.6
|
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
| chr8_+_104340594 | 0.62 |
ENSMUST00000034343.4
|
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr5_+_143651222 | 0.61 |
ENSMUST00000110727.1
|
Cyth3
|
cytohesin 3 |
| chr17_+_8849974 | 0.61 |
ENSMUST00000115720.1
|
Pde10a
|
phosphodiesterase 10A |
| chr19_-_46039621 | 0.61 |
ENSMUST00000056931.7
|
Ldb1
|
LIM domain binding 1 |
| chr3_-_52104891 | 0.61 |
ENSMUST00000121440.1
|
Maml3
|
mastermind like 3 (Drosophila) |
| chrX_+_56731779 | 0.61 |
ENSMUST00000023854.3
ENSMUST00000114769.2 |
Fhl1
|
four and a half LIM domains 1 |
| chr19_-_46045194 | 0.60 |
ENSMUST00000156585.1
ENSMUST00000152946.1 |
Ldb1
|
LIM domain binding 1 |
| chr11_+_71749914 | 0.60 |
ENSMUST00000150531.1
|
Wscd1
|
WSC domain containing 1 |
| chr4_-_133874682 | 0.59 |
ENSMUST00000168974.2
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr1_-_149961230 | 0.59 |
ENSMUST00000070200.8
|
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr1_-_75278345 | 0.58 |
ENSMUST00000039534.4
|
Resp18
|
regulated endocrine-specific protein 18 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Epas1_Bcl3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 14.4 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 1.6 | 7.9 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 1.4 | 5.8 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 1.2 | 3.6 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) |
| 1.2 | 3.6 | GO:2001076 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.9 | 2.8 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.8 | 4.2 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.7 | 2.8 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.6 | 2.5 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.6 | 3.0 | GO:0031444 | regulation of twitch skeletal muscle contraction(GO:0014724) slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.6 | 2.9 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.6 | 2.3 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.6 | 1.7 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.6 | 2.8 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.5 | 12.6 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.5 | 3.2 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.5 | 1.5 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.5 | 4.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.5 | 4.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.4 | 1.3 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.4 | 1.7 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.4 | 4.4 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.4 | 1.9 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.4 | 1.8 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.3 | 3.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.3 | 1.0 | GO:0070846 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.3 | 5.8 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.3 | 1.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.3 | 2.4 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.3 | 1.5 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.3 | 1.5 | GO:0071105 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.3 | 13.5 | GO:0032094 | response to food(GO:0032094) |
| 0.3 | 1.2 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.3 | 1.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 | 2.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.3 | 3.7 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.3 | 1.3 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.2 | 3.0 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.2 | 1.5 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.2 | 0.7 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.2 | 1.2 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.2 | 0.9 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 0.9 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 0.7 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.2 | 0.9 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.2 | 2.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 0.6 | GO:0003032 | detection of oxygen(GO:0003032) negative regulation of macrophage cytokine production(GO:0010936) |
| 0.2 | 1.0 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.2 | 1.0 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.2 | 1.0 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.2 | 3.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.2 | 2.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 0.8 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.2 | 0.2 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.2 | 1.9 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.2 | 0.5 | GO:0052564 | response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.2 | 0.7 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.9 | GO:1902608 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.2 | 0.5 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.2 | 3.6 | GO:0046685 | response to activity(GO:0014823) response to arsenic-containing substance(GO:0046685) |
| 0.2 | 0.5 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.2 | 0.3 | GO:0097048 | dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.1 | 10.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 1.0 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.1 | 0.7 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 4.0 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 2.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 2.6 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.7 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.5 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.1 | 0.9 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.1 | 1.3 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.1 | 1.2 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.1 | 1.6 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.4 | GO:0015819 | lysine transport(GO:0015819) |
| 0.1 | 1.1 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.1 | 0.9 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.8 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.6 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 1.5 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.1 | 0.4 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.1 | 0.6 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 1.3 | GO:0010919 | regulation of inositol phosphate biosynthetic process(GO:0010919) |
| 0.1 | 0.8 | GO:1904378 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 1.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.4 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 0.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.7 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 2.0 | GO:0045953 | negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.1 | 0.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 2.9 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.1 | 0.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.2 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 0.6 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.2 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.1 | 0.6 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.1 | 1.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 2.7 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.2 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) |
| 0.1 | 0.9 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.1 | 0.9 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.3 | GO:1903375 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) trigeminal ganglion development(GO:0061551) facioacoustic ganglion development(GO:1903375) |
| 0.1 | 0.7 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 0.6 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 2.0 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 1.1 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.1 | GO:0003274 | endocardial cushion fusion(GO:0003274) positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.5 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.9 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 1.4 | GO:0043267 | negative regulation of potassium ion transport(GO:0043267) |
| 0.0 | 0.3 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.8 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.5 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 1.1 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.6 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.2 | GO:0071351 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 0.0 | 0.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.4 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.4 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 7.2 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 2.7 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.0 | 0.2 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.3 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.7 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.5 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.4 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 1.1 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.9 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.9 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 1.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.8 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.6 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 2.3 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 1.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 2.9 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 2.7 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 1.3 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.0 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.5 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 1.2 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.5 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.0 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.5 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 3.1 | GO:0032496 | response to lipopolysaccharide(GO:0032496) |
| 0.0 | 1.6 | GO:0006165 | nucleoside diphosphate phosphorylation(GO:0006165) |
| 0.0 | 0.9 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 1.8 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 1.0 | GO:0017148 | negative regulation of translation(GO:0017148) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.4 | 4.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 1.1 | 5.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.8 | 5.8 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.7 | 2.8 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.4 | 1.2 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.4 | 1.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 1.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.3 | 1.0 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.2 | 0.7 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.2 | 1.0 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 3.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 6.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 1.7 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 0.5 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.1 | 3.0 | GO:0005859 | muscle myosin complex(GO:0005859) myosin filament(GO:0032982) |
| 0.1 | 0.8 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 4.7 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 3.1 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.6 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 1.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 2.3 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 2.0 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 1.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.2 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.1 | 3.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 4.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.2 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.0 | 0.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 2.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.4 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 4.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.5 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 2.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 3.0 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 7.7 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 3.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 3.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 3.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 9.2 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.5 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 3.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.9 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.3 | GO:0033643 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.0 | 0.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.9 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 4.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 12.6 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 2.4 | 9.8 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 1.1 | 3.2 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.7 | 5.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.7 | 2.9 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.5 | 4.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.5 | 4.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.5 | 14.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.4 | 5.3 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.4 | 1.2 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) glycoprotein transporter activity(GO:0034437) |
| 0.4 | 1.5 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.4 | 1.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.3 | 0.9 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.3 | 1.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 1.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 1.0 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.2 | 0.7 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.2 | 0.9 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.2 | 2.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 0.8 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 1.3 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.2 | 3.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 0.7 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 5.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 0.9 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.2 | 1.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 0.5 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.2 | 3.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 3.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 3.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.7 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 8.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.4 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 3.8 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.7 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.1 | 1.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.9 | GO:1990380 | Lys63-specific deubiquitinase activity(GO:0061578) Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.8 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.9 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 3.4 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.1 | 1.0 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 1.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.4 | GO:0015189 | L-ornithine transmembrane transporter activity(GO:0000064) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 1.9 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 1.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.2 | GO:0071820 | N-box binding(GO:0071820) |
| 0.1 | 0.5 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.2 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 5.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 3.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.7 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 1.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 0.4 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 2.8 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.1 | 1.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.2 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 2.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 2.9 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 2.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.7 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 2.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 3.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.2 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 1.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 3.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 2.0 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.5 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
| 0.0 | 1.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 1.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 3.1 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 1.7 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 7.0 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.5 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.0 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 7.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 2.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 1.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 4.9 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 6.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 4.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.8 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 7.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 4.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.8 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 1.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.9 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 1.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.7 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 14.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.3 | 11.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.2 | 4.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 3.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 4.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 3.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 4.5 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 1.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 2.6 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 3.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 2.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 3.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 0.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 2.2 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 0.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 1.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 2.0 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.7 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 1.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 1.9 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 1.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.8 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.2 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |