Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
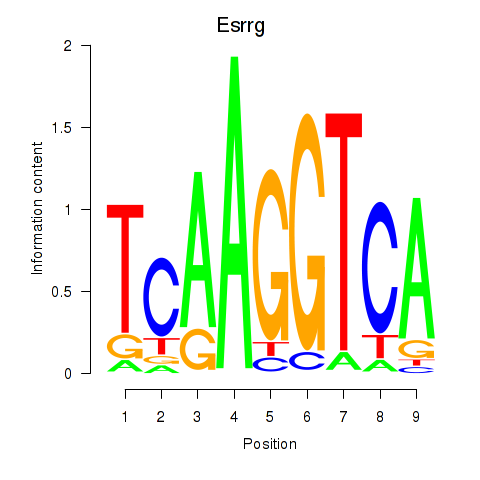
Results for Esrrg
Z-value: 0.63
Transcription factors associated with Esrrg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Esrrg
|
ENSMUSG00000026610.7 | estrogen-related receptor gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Esrrg | mm10_v2_chr1_+_187997835_187997882 | 0.51 | 1.6e-03 | Click! |
Activity profile of Esrrg motif
Sorted Z-values of Esrrg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_43524462 | 4.92 |
ENSMUST00000026196.7
|
Got1
|
glutamate oxaloacetate transaminase 1, soluble |
| chr4_-_107307118 | 4.61 |
ENSMUST00000126291.1
ENSMUST00000106748.1 ENSMUST00000129138.1 ENSMUST00000082426.3 |
Dio1
|
deiodinase, iodothyronine, type I |
| chr18_+_45268876 | 4.50 |
ENSMUST00000183850.1
ENSMUST00000066890.7 |
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr9_+_55326913 | 2.68 |
ENSMUST00000085754.3
ENSMUST00000034862.4 |
AI118078
|
expressed sequence AI118078 |
| chr13_+_84222286 | 1.85 |
ENSMUST00000057495.8
|
Tmem161b
|
transmembrane protein 161B |
| chr17_-_32420965 | 1.81 |
ENSMUST00000170392.1
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr2_-_25501717 | 1.62 |
ENSMUST00000015227.3
|
C8g
|
complement component 8, gamma polypeptide |
| chr5_-_92328068 | 1.53 |
ENSMUST00000113093.3
|
Cxcl9
|
chemokine (C-X-C motif) ligand 9 |
| chr9_-_107668967 | 1.35 |
ENSMUST00000177567.1
|
Slc38a3
|
solute carrier family 38, member 3 |
| chrX_-_72656135 | 1.33 |
ENSMUST00000055966.6
|
Gabra3
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 3 |
| chr4_-_63662910 | 1.33 |
ENSMUST00000184252.1
|
Gm11214
|
predicted gene 11214 |
| chr9_-_15301555 | 1.30 |
ENSMUST00000034414.8
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr7_-_81706905 | 1.22 |
ENSMUST00000026922.7
|
Homer2
|
homer homolog 2 (Drosophila) |
| chr19_-_4839286 | 1.06 |
ENSMUST00000037246.5
|
Ccs
|
copper chaperone for superoxide dismutase |
| chr13_+_9276477 | 0.96 |
ENSMUST00000174552.1
|
Dip2c
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr14_+_78849171 | 0.95 |
ENSMUST00000040990.5
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr3_-_90514250 | 0.95 |
ENSMUST00000107340.1
ENSMUST00000060738.8 |
S100a1
|
S100 calcium binding protein A1 |
| chr2_-_77170534 | 0.93 |
ENSMUST00000111833.2
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr2_+_152105722 | 0.91 |
ENSMUST00000099225.2
|
Srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr4_+_140961203 | 0.90 |
ENSMUST00000010007.8
|
Sdhb
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr8_-_84044982 | 0.89 |
ENSMUST00000061923.4
|
Rln3
|
relaxin 3 |
| chr4_-_19708922 | 0.88 |
ENSMUST00000108246.2
|
Wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr16_+_22951072 | 0.86 |
ENSMUST00000023590.8
|
Hrg
|
histidine-rich glycoprotein |
| chr1_-_136260873 | 0.86 |
ENSMUST00000086395.5
|
Gpr25
|
G protein-coupled receptor 25 |
| chr15_+_23036449 | 0.84 |
ENSMUST00000164787.1
|
Cdh18
|
cadherin 18 |
| chr15_-_76199835 | 0.81 |
ENSMUST00000054449.7
ENSMUST00000169714.1 ENSMUST00000165453.1 |
Plec
|
plectin |
| chr3_+_32736990 | 0.80 |
ENSMUST00000127477.1
ENSMUST00000121778.1 ENSMUST00000154257.1 |
Ndufb5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5 |
| chr2_-_73911323 | 0.77 |
ENSMUST00000111996.1
ENSMUST00000018914.2 |
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) |
| chr1_-_175625580 | 0.77 |
ENSMUST00000027810.7
|
Fh1
|
fumarate hydratase 1 |
| chr2_+_71981184 | 0.77 |
ENSMUST00000090826.5
ENSMUST00000102698.3 |
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr4_-_82505274 | 0.76 |
ENSMUST00000050872.8
ENSMUST00000064770.2 |
Nfib
|
nuclear factor I/B |
| chr7_-_4522427 | 0.75 |
ENSMUST00000098859.3
|
Tnni3
|
troponin I, cardiac 3 |
| chr6_+_91157373 | 0.75 |
ENSMUST00000155007.1
|
Hdac11
|
histone deacetylase 11 |
| chr2_+_145785980 | 0.74 |
ENSMUST00000110005.1
ENSMUST00000094480.4 |
Rin2
|
Ras and Rab interactor 2 |
| chr8_-_13494479 | 0.73 |
ENSMUST00000033828.5
|
Gas6
|
growth arrest specific 6 |
| chr7_-_17027807 | 0.71 |
ENSMUST00000142597.1
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr11_-_97573929 | 0.71 |
ENSMUST00000126287.1
ENSMUST00000107590.1 |
Srcin1
|
SRC kinase signaling inhibitor 1 |
| chr1_+_191906743 | 0.71 |
ENSMUST00000044954.6
|
Slc30a1
|
solute carrier family 30 (zinc transporter), member 1 |
| chr4_-_82505707 | 0.70 |
ENSMUST00000107248.1
ENSMUST00000107247.1 |
Nfib
|
nuclear factor I/B |
| chr2_+_140170641 | 0.67 |
ENSMUST00000044825.4
|
Ndufaf5
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 5 |
| chr8_+_94838321 | 0.67 |
ENSMUST00000034234.8
ENSMUST00000159871.1 |
Coq9
|
coenzyme Q9 homolog (yeast) |
| chr7_-_17027853 | 0.65 |
ENSMUST00000003183.5
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr10_+_128083273 | 0.64 |
ENSMUST00000026459.5
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr5_-_24995748 | 0.62 |
ENSMUST00000076306.5
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr4_-_129227883 | 0.60 |
ENSMUST00000106051.1
|
C77080
|
expressed sequence C77080 |
| chr19_-_34879452 | 0.60 |
ENSMUST00000036584.5
|
Pank1
|
pantothenate kinase 1 |
| chr1_-_138847579 | 0.59 |
ENSMUST00000093486.3
ENSMUST00000046870.6 |
Lhx9
|
LIM homeobox protein 9 |
| chr4_+_128993224 | 0.58 |
ENSMUST00000030583.6
ENSMUST00000102604.4 |
Ak2
|
adenylate kinase 2 |
| chr5_+_63649335 | 0.58 |
ENSMUST00000159584.1
|
3110047P20Rik
|
RIKEN cDNA 3110047P20 gene |
| chr9_-_63399216 | 0.57 |
ENSMUST00000168665.1
|
2300009A05Rik
|
RIKEN cDNA 2300009A05 gene |
| chr4_+_43632185 | 0.57 |
ENSMUST00000107874.2
|
Npr2
|
natriuretic peptide receptor 2 |
| chr11_-_53430779 | 0.57 |
ENSMUST00000061326.4
ENSMUST00000109021.3 |
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr7_+_83631959 | 0.56 |
ENSMUST00000075418.7
ENSMUST00000117410.1 |
Stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr3_+_90514435 | 0.55 |
ENSMUST00000048138.6
ENSMUST00000181271.1 |
S100a13
|
S100 calcium binding protein A13 |
| chr9_-_50603792 | 0.55 |
ENSMUST00000000175.4
|
Sdhd
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chr2_-_84678051 | 0.55 |
ENSMUST00000053664.8
ENSMUST00000111664.1 |
Tmx2
|
thioredoxin-related transmembrane protein 2 |
| chr16_+_45158725 | 0.53 |
ENSMUST00000023343.3
|
Atg3
|
autophagy related 3 |
| chrX_-_75382291 | 0.53 |
ENSMUST00000151772.1
ENSMUST00000033539.6 |
F8
|
coagulation factor VIII |
| chr11_-_94677404 | 0.53 |
ENSMUST00000116349.2
|
Xylt2
|
xylosyltransferase II |
| chr9_+_37401897 | 0.53 |
ENSMUST00000115048.1
|
Robo4
|
roundabout homolog 4 (Drosophila) |
| chr16_-_95459245 | 0.53 |
ENSMUST00000176345.1
ENSMUST00000121809.2 ENSMUST00000118113.1 ENSMUST00000122199.1 |
Erg
|
avian erythroblastosis virus E-26 (v-ets) oncogene related |
| chrX_+_160768013 | 0.52 |
ENSMUST00000033650.7
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr9_-_44980390 | 0.51 |
ENSMUST00000002101.5
ENSMUST00000160886.1 |
Cd3g
|
CD3 antigen, gamma polypeptide |
| chr10_+_79716588 | 0.51 |
ENSMUST00000099513.1
ENSMUST00000020581.2 |
Hcn2
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 2 |
| chr4_+_138434647 | 0.51 |
ENSMUST00000044058.4
ENSMUST00000105813.1 ENSMUST00000105815.1 |
Mul1
|
mitochondrial ubiquitin ligase activator of NFKB 1 |
| chr16_-_45158650 | 0.50 |
ENSMUST00000023344.3
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr6_-_113531575 | 0.49 |
ENSMUST00000032425.5
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr19_-_43674844 | 0.48 |
ENSMUST00000046038.7
|
Slc25a28
|
solute carrier family 25, member 28 |
| chr11_+_70844745 | 0.48 |
ENSMUST00000076270.6
ENSMUST00000179114.1 ENSMUST00000100928.4 ENSMUST00000177731.1 ENSMUST00000108533.3 ENSMUST00000081362.6 ENSMUST00000178245.1 |
Rabep1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr14_-_36919314 | 0.48 |
ENSMUST00000182797.1
|
Ccser2
|
coiled-coil serine rich 2 |
| chr19_-_6980420 | 0.48 |
ENSMUST00000070878.8
ENSMUST00000177752.1 |
Fkbp2
|
FK506 binding protein 2 |
| chr7_-_4445181 | 0.48 |
ENSMUST00000138798.1
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr4_-_108780503 | 0.47 |
ENSMUST00000106658.1
|
Zfyve9
|
zinc finger, FYVE domain containing 9 |
| chr9_+_78175898 | 0.46 |
ENSMUST00000180974.1
|
C920006O11Rik
|
RIKEN cDNA C920006O11 gene |
| chr10_-_43540945 | 0.46 |
ENSMUST00000147196.1
ENSMUST00000019932.3 |
1700021F05Rik
|
RIKEN cDNA 1700021F05 gene |
| chr2_+_130576170 | 0.46 |
ENSMUST00000028764.5
|
Oxt
|
oxytocin |
| chr14_-_36919513 | 0.45 |
ENSMUST00000182042.1
|
Ccser2
|
coiled-coil serine rich 2 |
| chr1_+_104768510 | 0.44 |
ENSMUST00000062528.8
|
Cdh20
|
cadherin 20 |
| chr5_+_122101146 | 0.43 |
ENSMUST00000147178.1
|
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr18_-_36744518 | 0.43 |
ENSMUST00000014438.4
|
Ndufa2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 |
| chr15_+_25742314 | 0.43 |
ENSMUST00000135981.1
|
Myo10
|
myosin X |
| chr4_-_128962420 | 0.42 |
ENSMUST00000119354.1
ENSMUST00000106068.1 ENSMUST00000030581.3 |
Adc
|
arginine decarboxylase |
| chr7_-_37772868 | 0.41 |
ENSMUST00000176205.1
|
Zfp536
|
zinc finger protein 536 |
| chr9_-_106891401 | 0.41 |
ENSMUST00000069036.7
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr7_-_4522794 | 0.40 |
ENSMUST00000140424.1
|
Tnni3
|
troponin I, cardiac 3 |
| chr15_+_76343504 | 0.40 |
ENSMUST00000023210.6
|
Cyc1
|
cytochrome c-1 |
| chr3_+_85574109 | 0.40 |
ENSMUST00000127348.1
ENSMUST00000107672.1 ENSMUST00000107674.1 |
Pet112
|
PET112 homolog (S. cerevisiae) |
| chr4_+_141115660 | 0.39 |
ENSMUST00000181450.1
|
4921514A10Rik
|
RIKEN cDNA 4921514A10 gene |
| chr19_+_6996114 | 0.39 |
ENSMUST00000088223.5
|
Trpt1
|
tRNA phosphotransferase 1 |
| chr1_-_131279544 | 0.39 |
ENSMUST00000062108.3
|
Ikbke
|
inhibitor of kappaB kinase epsilon |
| chr2_-_103073335 | 0.38 |
ENSMUST00000132449.1
ENSMUST00000111183.1 ENSMUST00000011058.2 |
Pdhx
|
pyruvate dehydrogenase complex, component X |
| chr16_+_4684070 | 0.38 |
ENSMUST00000060067.5
ENSMUST00000115854.3 |
Dnaja3
|
DnaJ (Hsp40) homolog, subfamily A, member 3 |
| chr2_+_155517948 | 0.38 |
ENSMUST00000029135.8
ENSMUST00000065973.2 ENSMUST00000103142.5 |
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr5_+_102768771 | 0.38 |
ENSMUST00000112852.1
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr11_-_84870188 | 0.38 |
ENSMUST00000154915.2
|
Ggnbp2
|
gametogenetin binding protein 2 |
| chr9_+_37401993 | 0.38 |
ENSMUST00000115046.1
ENSMUST00000102895.4 |
Robo4
|
roundabout homolog 4 (Drosophila) |
| chr11_-_33276334 | 0.37 |
ENSMUST00000183831.1
|
Gm12117
|
predicted gene 12117 |
| chr11_+_93996082 | 0.37 |
ENSMUST00000041956.7
|
Spag9
|
sperm associated antigen 9 |
| chr19_-_47090610 | 0.37 |
ENSMUST00000096014.3
|
Usmg5
|
upregulated during skeletal muscle growth 5 |
| chrX_+_160768179 | 0.36 |
ENSMUST00000112368.2
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr19_-_57314896 | 0.36 |
ENSMUST00000111524.1
|
Ablim1
|
actin-binding LIM protein 1 |
| chr13_-_74350206 | 0.36 |
ENSMUST00000022062.7
|
Sdha
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr2_+_29965560 | 0.35 |
ENSMUST00000113717.1
ENSMUST00000113719.2 ENSMUST00000100225.2 ENSMUST00000113741.1 ENSMUST00000095083.4 ENSMUST00000046257.7 |
Sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr1_+_36471590 | 0.35 |
ENSMUST00000153128.1
|
Cnnm4
|
cyclin M4 |
| chr7_-_19665005 | 0.35 |
ENSMUST00000055242.9
|
Clptm1
|
cleft lip and palate associated transmembrane protein 1 |
| chr2_-_10080055 | 0.34 |
ENSMUST00000130067.1
ENSMUST00000139810.1 |
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chrX_+_56963325 | 0.34 |
ENSMUST00000096431.3
|
Gpr112
|
G protein-coupled receptor 112 |
| chr2_+_127587214 | 0.34 |
ENSMUST00000028852.6
|
Mrps5
|
mitochondrial ribosomal protein S5 |
| chr14_+_58076507 | 0.33 |
ENSMUST00000166770.1
|
Fgf9
|
fibroblast growth factor 9 |
| chr1_-_84696182 | 0.33 |
ENSMUST00000049126.6
|
Dner
|
delta/notch-like EGF-related receptor |
| chr2_-_77170592 | 0.33 |
ENSMUST00000164114.2
ENSMUST00000049544.7 |
Ccdc141
|
coiled-coil domain containing 141 |
| chr1_-_5070281 | 0.33 |
ENSMUST00000147158.1
ENSMUST00000118000.1 |
Rgs20
|
regulator of G-protein signaling 20 |
| chr18_+_77773956 | 0.33 |
ENSMUST00000114748.1
|
Atp5a1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 |
| chr1_-_191907527 | 0.32 |
ENSMUST00000069573.5
|
1700034H15Rik
|
RIKEN cDNA 1700034H15 gene |
| chr7_+_120635176 | 0.32 |
ENSMUST00000033176.5
|
Uqcrc2
|
ubiquinol cytochrome c reductase core protein 2 |
| chrX_-_136976859 | 0.32 |
ENSMUST00000127404.1
ENSMUST00000113071.1 |
Tmsb15l
Tmsb15b1
|
thymosin beta 15b like thymosin beta 15b1 |
| chr4_-_82505749 | 0.32 |
ENSMUST00000107245.2
ENSMUST00000107246.1 |
Nfib
|
nuclear factor I/B |
| chr9_+_50603892 | 0.32 |
ENSMUST00000044051.4
|
Timm8b
|
translocase of inner mitochondrial membrane 8B |
| chr18_-_34954302 | 0.31 |
ENSMUST00000025217.8
|
Hspa9
|
heat shock protein 9 |
| chr5_-_31291026 | 0.30 |
ENSMUST00000041565.7
|
Ift172
|
intraflagellar transport 172 |
| chr3_+_146450467 | 0.30 |
ENSMUST00000061937.6
ENSMUST00000029840.3 |
Ctbs
|
chitobiase, di-N-acetyl- |
| chr19_-_6996025 | 0.30 |
ENSMUST00000041686.3
ENSMUST00000180765.1 |
Nudt22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
| chr4_-_108780782 | 0.29 |
ENSMUST00000106657.1
|
Zfyve9
|
zinc finger, FYVE domain containing 9 |
| chr5_-_134456702 | 0.29 |
ENSMUST00000073161.5
ENSMUST00000171794.2 ENSMUST00000111245.2 ENSMUST00000100654.3 ENSMUST00000167084.2 ENSMUST00000100652.3 ENSMUST00000100650.3 ENSMUST00000074114.5 |
Gtf2ird1
|
general transcription factor II I repeat domain-containing 1 |
| chr17_+_47385386 | 0.29 |
ENSMUST00000024774.7
ENSMUST00000145462.1 |
Guca1b
|
guanylate cyclase activator 1B |
| chr14_+_25694170 | 0.29 |
ENSMUST00000022419.6
|
Ppif
|
peptidylprolyl isomerase F (cyclophilin F) |
| chr5_-_49285653 | 0.29 |
ENSMUST00000175660.1
|
Kcnip4
|
Kv channel interacting protein 4 |
| chr11_+_94044331 | 0.29 |
ENSMUST00000024979.8
|
Spag9
|
sperm associated antigen 9 |
| chr12_-_71136611 | 0.27 |
ENSMUST00000021486.8
ENSMUST00000166120.1 |
Timm9
|
translocase of inner mitochondrial membrane 9 |
| chr2_-_121806988 | 0.27 |
ENSMUST00000110592.1
|
Frmd5
|
FERM domain containing 5 |
| chr12_-_103631404 | 0.27 |
ENSMUST00000121625.1
ENSMUST00000044231.5 |
Serpina10
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr11_-_102407315 | 0.26 |
ENSMUST00000149777.1
ENSMUST00000154001.1 |
Slc25a39
|
solute carrier family 25, member 39 |
| chr16_+_21891969 | 0.26 |
ENSMUST00000042065.6
|
Map3k13
|
mitogen-activated protein kinase kinase kinase 13 |
| chrX_-_71492592 | 0.26 |
ENSMUST00000080035.4
|
Cd99l2
|
CD99 antigen-like 2 |
| chr2_-_121807024 | 0.26 |
ENSMUST00000138157.1
|
Frmd5
|
FERM domain containing 5 |
| chr7_-_4445637 | 0.25 |
ENSMUST00000008579.7
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr16_+_10545339 | 0.25 |
ENSMUST00000066345.7
ENSMUST00000115824.3 ENSMUST00000155633.1 |
Clec16a
|
C-type lectin domain family 16, member A |
| chr14_-_20794009 | 0.25 |
ENSMUST00000100837.3
ENSMUST00000080440.6 ENSMUST00000071816.6 |
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr3_-_32737147 | 0.25 |
ENSMUST00000043966.7
|
Mrpl47
|
mitochondrial ribosomal protein L47 |
| chr8_-_94838255 | 0.25 |
ENSMUST00000161762.1
ENSMUST00000162538.1 |
Ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chr13_-_21501418 | 0.25 |
ENSMUST00000044043.2
|
Gm11273
|
predicted gene 11273 |
| chr14_+_32085804 | 0.24 |
ENSMUST00000170600.1
ENSMUST00000168986.1 ENSMUST00000169649.1 |
Oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr15_-_35938009 | 0.23 |
ENSMUST00000156915.1
|
Cox6c
|
cytochrome c oxidase subunit VIc |
| chr11_-_84870646 | 0.22 |
ENSMUST00000018547.2
|
Ggnbp2
|
gametogenetin binding protein 2 |
| chr3_-_107517321 | 0.22 |
ENSMUST00000166892.1
|
Slc6a17
|
solute carrier family 6 (neurotransmitter transporter), member 17 |
| chr5_+_145204523 | 0.22 |
ENSMUST00000085671.3
ENSMUST00000031601.7 |
Zkscan5
|
zinc finger with KRAB and SCAN domains 5 |
| chr7_-_116237767 | 0.22 |
ENSMUST00000182834.1
|
Plekha7
|
pleckstrin homology domain containing, family A member 7 |
| chr5_+_118065360 | 0.22 |
ENSMUST00000031305.3
|
Gm9754
|
predicted gene 9754 |
| chr7_-_126475082 | 0.22 |
ENSMUST00000032978.6
|
Sh2b1
|
SH2B adaptor protein 1 |
| chr11_+_74649462 | 0.21 |
ENSMUST00000092915.5
ENSMUST00000117818.1 |
Cluh
|
clustered mitochondria (cluA/CLU1) homolog |
| chr16_-_91931643 | 0.21 |
ENSMUST00000023677.3
|
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr4_-_40279389 | 0.21 |
ENSMUST00000108108.2
ENSMUST00000095128.3 |
Ndufb6
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6 |
| chr17_+_84957993 | 0.21 |
ENSMUST00000080217.6
ENSMUST00000112304.2 |
Ppm1b
|
protein phosphatase 1B, magnesium dependent, beta isoform |
| chr6_-_125165576 | 0.20 |
ENSMUST00000183272.1
ENSMUST00000182052.1 ENSMUST00000182277.1 |
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr10_-_84440591 | 0.20 |
ENSMUST00000020220.8
|
Nuak1
|
NUAK family, SNF1-like kinase, 1 |
| chr1_+_75546522 | 0.20 |
ENSMUST00000138814.1
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr18_-_43438280 | 0.19 |
ENSMUST00000121805.1
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr18_+_36744656 | 0.19 |
ENSMUST00000007042.5
|
Ik
|
IK cytokine |
| chr13_+_96542727 | 0.18 |
ENSMUST00000077672.4
ENSMUST00000109444.2 |
Col4a3bp
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein |
| chr5_-_137613759 | 0.18 |
ENSMUST00000155251.1
ENSMUST00000124693.1 |
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr5_-_92348871 | 0.18 |
ENSMUST00000038816.6
ENSMUST00000118006.1 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr10_+_80148263 | 0.18 |
ENSMUST00000099492.3
ENSMUST00000042057.5 |
Midn
|
midnolin |
| chrX_-_8018492 | 0.18 |
ENSMUST00000033503.2
|
Glod5
|
glyoxalase domain containing 5 |
| chr11_-_84870712 | 0.17 |
ENSMUST00000170741.1
ENSMUST00000172405.1 ENSMUST00000100686.3 ENSMUST00000108081.2 |
Ggnbp2
|
gametogenetin binding protein 2 |
| chr9_+_54586450 | 0.17 |
ENSMUST00000167866.1
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr2_+_178119166 | 0.17 |
ENSMUST00000108916.1
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr11_+_69088490 | 0.16 |
ENSMUST00000021273.6
ENSMUST00000117780.1 |
Vamp2
|
vesicle-associated membrane protein 2 |
| chr11_-_115699307 | 0.16 |
ENSMUST00000106499.1
|
Grb2
|
growth factor receptor bound protein 2 |
| chr1_+_36691487 | 0.15 |
ENSMUST00000081180.4
|
Cox5b
|
cytochrome c oxidase subunit Vb |
| chrX_-_71492799 | 0.15 |
ENSMUST00000037391.5
ENSMUST00000114586.2 ENSMUST00000114587.2 |
Cd99l2
|
CD99 antigen-like 2 |
| chr4_+_99955715 | 0.15 |
ENSMUST00000102783.4
|
Pgm2
|
phosphoglucomutase 2 |
| chr2_+_32450444 | 0.15 |
ENSMUST00000048431.2
|
Naif1
|
nuclear apoptosis inducing factor 1 |
| chr16_+_10545390 | 0.15 |
ENSMUST00000115827.1
ENSMUST00000038145.6 ENSMUST00000150894.1 |
Clec16a
|
C-type lectin domain family 16, member A |
| chr14_+_21500879 | 0.15 |
ENSMUST00000182964.1
|
Kat6b
|
K(lysine) acetyltransferase 6B |
| chr1_-_131097535 | 0.15 |
ENSMUST00000016672.4
|
Mapkapk2
|
MAP kinase-activated protein kinase 2 |
| chrX_-_75382615 | 0.14 |
ENSMUST00000147349.1
|
F8
|
coagulation factor VIII |
| chr11_+_76407143 | 0.14 |
ENSMUST00000021203.6
ENSMUST00000152183.1 |
Timm22
|
translocase of inner mitochondrial membrane 22 |
| chr5_-_121527186 | 0.13 |
ENSMUST00000152270.1
|
Mapkapk5
|
MAP kinase-activated protein kinase 5 |
| chr6_-_92183972 | 0.13 |
ENSMUST00000140438.1
|
Mrps25
|
mitochondrial ribosomal protein S25 |
| chr17_+_26123514 | 0.13 |
ENSMUST00000025014.8
|
Mrpl28
|
mitochondrial ribosomal protein L28 |
| chr17_-_29347902 | 0.12 |
ENSMUST00000095427.4
ENSMUST00000118366.1 |
Mtch1
|
mitochondrial carrier homolog 1 (C. elegans) |
| chr5_-_24445166 | 0.12 |
ENSMUST00000115043.1
ENSMUST00000115041.1 |
Fastk
|
Fas-activated serine/threonine kinase |
| chr9_-_54661666 | 0.11 |
ENSMUST00000128624.1
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr9_-_54647199 | 0.11 |
ENSMUST00000128163.1
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr11_-_84870812 | 0.11 |
ENSMUST00000168434.1
|
Ggnbp2
|
gametogenetin binding protein 2 |
| chr13_-_91876876 | 0.10 |
ENSMUST00000022122.3
|
Ckmt2
|
creatine kinase, mitochondrial 2 |
| chr1_+_131910458 | 0.10 |
ENSMUST00000062264.6
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr2_+_178118975 | 0.10 |
ENSMUST00000108917.1
|
Phactr3
|
phosphatase and actin regulator 3 |
| chrX_+_105764262 | 0.10 |
ENSMUST00000033581.3
|
Fgf16
|
fibroblast growth factor 16 |
| chr7_+_128129536 | 0.10 |
ENSMUST00000033053.6
|
Itgax
|
integrin alpha X |
| chr18_-_77186257 | 0.09 |
ENSMUST00000097520.2
|
Gm7276
|
predicted gene 7276 |
| chr1_+_6734827 | 0.09 |
ENSMUST00000139838.1
|
St18
|
suppression of tumorigenicity 18 |
| chr6_-_71823805 | 0.09 |
ENSMUST00000065103.2
|
Mrpl35
|
mitochondrial ribosomal protein L35 |
| chr18_+_77185815 | 0.08 |
ENSMUST00000079618.4
|
St8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr6_+_83349446 | 0.08 |
ENSMUST00000136501.1
|
Bola3
|
bolA-like 3 (E. coli) |
| chr11_+_70764209 | 0.08 |
ENSMUST00000060444.5
|
Zfp3
|
zinc finger protein 3 |
| chr2_-_112480817 | 0.07 |
ENSMUST00000099589.2
|
Chrm5
|
cholinergic receptor, muscarinic 5 |
| chr3_-_107555069 | 0.07 |
ENSMUST00000052853.6
|
Ubl4b
|
ubiquitin-like 4B |
| chr2_+_130012336 | 0.07 |
ENSMUST00000110299.2
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr1_+_75435930 | 0.06 |
ENSMUST00000037796.7
ENSMUST00000113584.1 ENSMUST00000145166.1 ENSMUST00000143730.1 ENSMUST00000133418.1 ENSMUST00000144874.1 ENSMUST00000140287.1 |
Gmppa
|
GDP-mannose pyrophosphorylase A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Esrrg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.6 | 4.5 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.5 | 1.4 | GO:2000485 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) regulation of glutamine transport(GO:2000485) |
| 0.4 | 1.8 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.4 | 1.8 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.3 | 0.9 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.3 | 1.6 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.2 | 0.7 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.2 | 0.9 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.2 | 0.9 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.2 | 0.8 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.2 | 0.7 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) |
| 0.2 | 0.9 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.2 | 1.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.8 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.5 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 0.6 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 1.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.6 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 0.3 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 | 0.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.4 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.1 | 0.6 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 0.5 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.8 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.1 | 0.5 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.7 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.6 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 0.4 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.9 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.1 | 0.6 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.1 | 0.3 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.1 | 0.5 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.3 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 1.7 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.1 | 0.9 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 0.4 | GO:0006083 | acetate metabolic process(GO:0006083) short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.2 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.1 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 1.2 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.1 | 0.6 | GO:1900194 | receptor guanylyl cyclase signaling pathway(GO:0007168) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.4 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 0.2 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.1 | 0.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.6 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.1 | 2.1 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.5 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 4.6 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.0 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.2 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 1.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.8 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.1 | GO:0070342 | brown fat cell proliferation(GO:0070342) regulation of brown fat cell proliferation(GO:0070347) |
| 0.0 | 0.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.3 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 1.3 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.5 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.3 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.7 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.2 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.2 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 1.1 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.0 | 0.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.4 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 0.6 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.7 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.9 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 | 0.1 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.3 | GO:2000398 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 | 0.2 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.3 | 1.3 | GO:0036019 | endolysosome(GO:0036019) |
| 0.2 | 1.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 0.5 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.2 | 1.3 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 0.8 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 1.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.6 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.1 | 0.7 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 1.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 4.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 1.3 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.1 | 1.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.3 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.1 | 1.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.4 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 1.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.8 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.5 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.4 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 5.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.2 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.6 | GO:0031901 | early endosome membrane(GO:0031901) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0070546 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 1.5 | 4.6 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 1.1 | 4.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.6 | 1.8 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.5 | 1.4 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.4 | 1.8 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.3 | 1.7 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.3 | 1.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 1.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 1.0 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.2 | 0.5 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.2 | 0.5 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.2 | 1.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.4 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.6 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 1.6 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.6 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 1.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.6 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.5 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.9 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.4 | GO:0008384 | NF-kappaB-inducing kinase activity(GO:0004704) IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.4 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.6 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.7 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 0.2 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.1 | 0.3 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.6 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 1.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.7 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 1.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.7 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.5 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 1.4 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.5 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0016933 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.2 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 3.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 2.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 0.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 1.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 3.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 5.0 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.0 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |