Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Etv4
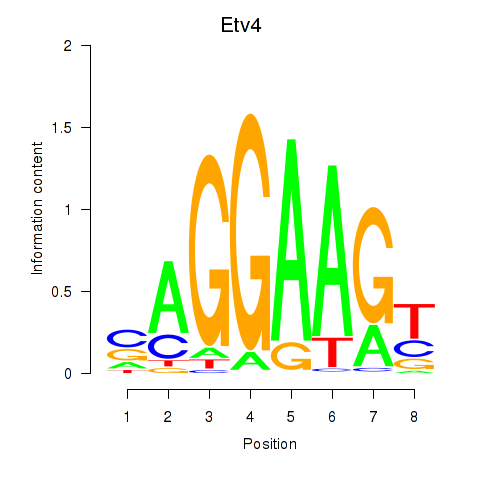
Z-value: 1.46
Transcription factors associated with Etv4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Etv4
|
ENSMUSG00000017724.8 | ets variant 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Etv4 | mm10_v2_chr11_-_101785252_101785371 | 0.79 | 1.2e-08 | Click! |
Activity profile of Etv4 motif
Sorted Z-values of Etv4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_87793470 | 19.07 |
ENSMUST00000020779.4
|
Mpo
|
myeloperoxidase |
| chr11_+_87793722 | 16.65 |
ENSMUST00000143021.2
|
Mpo
|
myeloperoxidase |
| chr7_+_24370255 | 10.25 |
ENSMUST00000171904.1
|
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr1_+_40515362 | 10.01 |
ENSMUST00000027237.5
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr2_-_28084877 | 9.36 |
ENSMUST00000028179.8
ENSMUST00000117486.1 ENSMUST00000135472.1 |
Fcnb
|
ficolin B |
| chr9_-_123968683 | 9.17 |
ENSMUST00000026911.4
|
Ccr1
|
chemokine (C-C motif) receptor 1 |
| chr11_-_102469839 | 8.38 |
ENSMUST00000103086.3
|
Itga2b
|
integrin alpha 2b |
| chrX_-_7964166 | 8.19 |
ENSMUST00000128449.1
|
Gata1
|
GATA binding protein 1 |
| chr8_+_72761868 | 7.34 |
ENSMUST00000058099.8
|
F2rl3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr7_-_127137807 | 7.24 |
ENSMUST00000049931.5
|
Spn
|
sialophorin |
| chr17_+_48359891 | 7.08 |
ENSMUST00000024792.6
|
Treml1
|
triggering receptor expressed on myeloid cells-like 1 |
| chr8_-_71723308 | 6.91 |
ENSMUST00000125092.1
|
Fcho1
|
FCH domain only 1 |
| chr13_-_110357136 | 6.80 |
ENSMUST00000058806.5
|
Gapt
|
Grb2-binding adaptor, transmembrane |
| chr2_+_173022360 | 6.79 |
ENSMUST00000173997.1
|
Rbm38
|
RNA binding motif protein 38 |
| chr14_+_80000292 | 6.78 |
ENSMUST00000088735.3
|
Olfm4
|
olfactomedin 4 |
| chr7_+_131032061 | 6.68 |
ENSMUST00000084509.3
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr3_-_106167564 | 6.60 |
ENSMUST00000063062.8
|
Chi3l3
|
chitinase 3-like 3 |
| chr4_-_118620763 | 6.48 |
ENSMUST00000071972.4
|
Wdr65
|
WD repeat domain 65 |
| chr8_-_85380964 | 6.30 |
ENSMUST00000122452.1
|
Mylk3
|
myosin light chain kinase 3 |
| chr17_+_33638056 | 6.05 |
ENSMUST00000052079.7
|
Pram1
|
PML-RAR alpha-regulated adaptor molecule 1 |
| chr19_-_7019423 | 5.67 |
ENSMUST00000040772.8
|
Fermt3
|
fermitin family homolog 3 (Drosophila) |
| chr11_-_72550255 | 5.66 |
ENSMUST00000021154.6
|
Spns3
|
spinster homolog 3 |
| chr6_-_40585783 | 5.42 |
ENSMUST00000177178.1
ENSMUST00000129948.2 ENSMUST00000101491.4 |
Clec5a
|
C-type lectin domain family 5, member a |
| chr3_+_103832562 | 5.37 |
ENSMUST00000062945.5
|
Bcl2l15
|
BCLl2-like 15 |
| chr7_+_127091426 | 5.32 |
ENSMUST00000056288.5
|
AI467606
|
expressed sequence AI467606 |
| chr1_+_174041933 | 5.04 |
ENSMUST00000052975.4
|
Olfr433
|
olfactory receptor 433 |
| chr13_-_37049203 | 5.01 |
ENSMUST00000037491.8
|
F13a1
|
coagulation factor XIII, A1 subunit |
| chr17_-_32403551 | 5.00 |
ENSMUST00000135618.1
ENSMUST00000063824.7 |
Rasal3
|
RAS protein activator like 3 |
| chr2_-_32387760 | 4.83 |
ENSMUST00000050785.8
|
Lcn2
|
lipocalin 2 |
| chr17_-_32403526 | 4.77 |
ENSMUST00000137458.1
|
Rasal3
|
RAS protein activator like 3 |
| chr7_-_127218390 | 4.72 |
ENSMUST00000142356.1
ENSMUST00000106314.1 |
Sept1
|
septin 1 |
| chr7_+_43437073 | 4.72 |
ENSMUST00000070518.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr15_+_84324716 | 4.71 |
ENSMUST00000023074.2
|
Parvg
|
parvin, gamma |
| chr11_+_11686213 | 4.67 |
ENSMUST00000076700.4
ENSMUST00000048122.6 |
Ikzf1
|
IKAROS family zinc finger 1 |
| chr11_+_103171081 | 4.66 |
ENSMUST00000042286.5
|
Fmnl1
|
formin-like 1 |
| chr11_+_11685909 | 4.65 |
ENSMUST00000065433.5
|
Ikzf1
|
IKAROS family zinc finger 1 |
| chr2_-_156839790 | 4.64 |
ENSMUST00000134838.1
ENSMUST00000137463.1 ENSMUST00000149275.2 |
Gm14230
|
predicted gene 14230 |
| chr7_-_126369543 | 4.62 |
ENSMUST00000032997.6
|
Lat
|
linker for activation of T cells |
| chr9_+_56089962 | 4.53 |
ENSMUST00000059206.7
|
Pstpip1
|
proline-serine-threonine phosphatase-interacting protein 1 |
| chr11_-_76509419 | 4.37 |
ENSMUST00000094012.4
|
Abr
|
active BCR-related gene |
| chr2_-_32083783 | 4.36 |
ENSMUST00000056406.6
|
Fam78a
|
family with sequence similarity 78, member A |
| chr2_-_164356067 | 4.33 |
ENSMUST00000165980.1
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr17_-_24527925 | 4.32 |
ENSMUST00000176652.1
|
Traf7
|
TNF receptor-associated factor 7 |
| chr14_+_65266701 | 4.29 |
ENSMUST00000169656.1
|
Fbxo16
|
F-box protein 16 |
| chr16_+_32608973 | 4.27 |
ENSMUST00000120680.1
|
Tfrc
|
transferrin receptor |
| chr9_+_65587187 | 4.23 |
ENSMUST00000047099.5
ENSMUST00000131483.1 ENSMUST00000141046.1 |
Pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr2_-_170406501 | 4.19 |
ENSMUST00000154650.1
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr17_-_24527830 | 4.14 |
ENSMUST00000176353.1
ENSMUST00000176237.1 |
Traf7
|
TNF receptor-associated factor 7 |
| chr4_-_152448808 | 4.07 |
ENSMUST00000159840.1
ENSMUST00000105648.2 |
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chrX_-_102251852 | 3.99 |
ENSMUST00000101336.3
ENSMUST00000136277.1 |
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chr3_-_14778452 | 3.98 |
ENSMUST00000094365.4
|
Car1
|
carbonic anhydrase 1 |
| chr11_+_115887601 | 3.90 |
ENSMUST00000167507.2
|
Myo15b
|
myosin XVB |
| chr6_-_123289862 | 3.89 |
ENSMUST00000032239.4
ENSMUST00000177367.1 |
Clec4e
|
C-type lectin domain family 4, member e |
| chr3_-_20242173 | 3.84 |
ENSMUST00000001921.1
|
Cpa3
|
carboxypeptidase A3, mast cell |
| chr3_+_28781305 | 3.83 |
ENSMUST00000060500.7
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr9_+_65587149 | 3.82 |
ENSMUST00000134538.1
ENSMUST00000136205.1 |
Pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr2_-_164356507 | 3.81 |
ENSMUST00000109367.3
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr8_-_46211284 | 3.81 |
ENSMUST00000034049.4
|
Slc25a4
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 |
| chr16_-_92826004 | 3.76 |
ENSMUST00000023673.7
|
Runx1
|
runt related transcription factor 1 |
| chr2_-_118728430 | 3.74 |
ENSMUST00000102524.1
|
Plcb2
|
phospholipase C, beta 2 |
| chr12_-_32208609 | 3.67 |
ENSMUST00000053215.7
|
Pik3cg
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide |
| chr1_+_171388954 | 3.66 |
ENSMUST00000056449.8
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr11_+_61126747 | 3.64 |
ENSMUST00000010286.1
ENSMUST00000146033.1 ENSMUST00000139422.1 |
Tnfrsf13b
|
tumor necrosis factor receptor superfamily, member 13b |
| chr1_+_152807877 | 3.61 |
ENSMUST00000027754.6
|
Ncf2
|
neutrophil cytosolic factor 2 |
| chr2_+_91650116 | 3.59 |
ENSMUST00000111331.2
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr7_-_100856289 | 3.58 |
ENSMUST00000139604.1
|
Relt
|
RELT tumor necrosis factor receptor |
| chr4_-_63403330 | 3.53 |
ENSMUST00000035724.4
|
Akna
|
AT-hook transcription factor |
| chr17_+_32403006 | 3.51 |
ENSMUST00000065921.5
|
A530088E08Rik
|
RIKEN cDNA A530088E08 gene |
| chr11_+_9191934 | 3.49 |
ENSMUST00000042740.6
|
Abca13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr7_-_127218303 | 3.48 |
ENSMUST00000106313.1
|
Sept1
|
septin 1 |
| chr7_+_24897381 | 3.46 |
ENSMUST00000003469.7
|
Cd79a
|
CD79A antigen (immunoglobulin-associated alpha) |
| chr17_+_34590162 | 3.44 |
ENSMUST00000173772.1
|
Gpsm3
|
G-protein signalling modulator 3 (AGS3-like, C. elegans) |
| chr16_+_32608920 | 3.43 |
ENSMUST00000023486.8
|
Tfrc
|
transferrin receptor |
| chr2_+_91650169 | 3.36 |
ENSMUST00000090614.4
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr17_+_34605855 | 3.36 |
ENSMUST00000037489.8
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| chr4_-_117182623 | 3.33 |
ENSMUST00000065896.2
|
Kif2c
|
kinesin family member 2C |
| chr17_+_34589799 | 3.32 |
ENSMUST00000038244.8
|
Gpsm3
|
G-protein signalling modulator 3 (AGS3-like, C. elegans) |
| chr11_-_59163281 | 3.31 |
ENSMUST00000069631.2
|
Iba57
|
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) |
| chr2_+_127336152 | 3.30 |
ENSMUST00000028846.6
|
Dusp2
|
dual specificity phosphatase 2 |
| chr1_-_75506331 | 3.29 |
ENSMUST00000113567.2
ENSMUST00000113565.2 |
Obsl1
|
obscurin-like 1 |
| chr14_-_76556662 | 3.29 |
ENSMUST00000064517.7
|
Serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr6_+_60944472 | 3.28 |
ENSMUST00000129603.1
|
Mmrn1
|
multimerin 1 |
| chr15_+_78244781 | 3.28 |
ENSMUST00000096357.5
ENSMUST00000133618.1 |
Ncf4
|
neutrophil cytosolic factor 4 |
| chr4_-_141078302 | 3.26 |
ENSMUST00000030760.8
|
Necap2
|
NECAP endocytosis associated 2 |
| chr6_+_145121727 | 3.22 |
ENSMUST00000032396.6
|
Lrmp
|
lymphoid-restricted membrane protein |
| chr14_-_33185489 | 3.21 |
ENSMUST00000159606.1
|
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr16_-_75909272 | 3.17 |
ENSMUST00000114239.2
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr2_-_26021679 | 3.17 |
ENSMUST00000036509.7
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr6_-_70792155 | 3.14 |
ENSMUST00000066134.5
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr17_+_35135463 | 3.13 |
ENSMUST00000173535.1
ENSMUST00000173952.1 |
Bag6
|
BCL2-associated athanogene 6 |
| chr3_+_103832741 | 3.13 |
ENSMUST00000106822.1
|
Bcl2l15
|
BCLl2-like 15 |
| chr12_-_8539545 | 3.12 |
ENSMUST00000095863.3
ENSMUST00000165657.1 |
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chr2_-_26021532 | 3.11 |
ENSMUST00000136750.1
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr2_-_73486456 | 3.08 |
ENSMUST00000141264.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr4_-_83021102 | 3.07 |
ENSMUST00000071708.5
|
Frem1
|
Fras1 related extracellular matrix protein 1 |
| chr6_+_113531675 | 3.04 |
ENSMUST00000036340.5
ENSMUST00000101051.2 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr1_+_135799833 | 2.98 |
ENSMUST00000148201.1
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chr4_-_129573637 | 2.96 |
ENSMUST00000102596.1
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr4_-_133872997 | 2.95 |
ENSMUST00000137486.2
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr16_-_3718105 | 2.91 |
ENSMUST00000023180.7
ENSMUST00000100222.2 |
Mefv
|
Mediterranean fever |
| chr2_-_26360873 | 2.89 |
ENSMUST00000028294.6
|
Card9
|
caspase recruitment domain family, member 9 |
| chr19_+_4154606 | 2.86 |
ENSMUST00000061086.8
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chr4_-_133872304 | 2.86 |
ENSMUST00000157067.2
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr5_+_134676490 | 2.85 |
ENSMUST00000100641.2
|
Gm10369
|
predicted gene 10369 |
| chr4_-_156228540 | 2.83 |
ENSMUST00000105571.2
|
Plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr3_-_109027600 | 2.82 |
ENSMUST00000171143.1
|
Fam102b
|
family with sequence similarity 102, member B |
| chr3_-_137981523 | 2.81 |
ENSMUST00000136613.1
ENSMUST00000029806.6 |
Dapp1
|
dual adaptor for phosphotyrosine and 3-phosphoinositides 1 |
| chr9_-_20952838 | 2.81 |
ENSMUST00000004202.9
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr11_-_6520894 | 2.77 |
ENSMUST00000003459.3
|
Myo1g
|
myosin IG |
| chr9_-_66126559 | 2.76 |
ENSMUST00000137542.1
|
Snx1
|
sorting nexin 1 |
| chr11_-_94653964 | 2.75 |
ENSMUST00000039949.4
|
Eme1
|
essential meiotic endonuclease 1 homolog 1 (S. pombe) |
| chr6_+_125552948 | 2.74 |
ENSMUST00000112254.1
ENSMUST00000112253.1 ENSMUST00000001995.7 |
Vwf
|
Von Willebrand factor homolog |
| chrX_-_8090442 | 2.74 |
ENSMUST00000033505.6
|
Was
|
Wiskott-Aldrich syndrome homolog (human) |
| chr4_+_135152496 | 2.73 |
ENSMUST00000119564.1
|
Runx3
|
runt related transcription factor 3 |
| chr1_+_87620334 | 2.72 |
ENSMUST00000042275.8
ENSMUST00000168783.1 |
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr17_+_35135196 | 2.72 |
ENSMUST00000172571.1
ENSMUST00000173491.1 |
Bag6
|
BCL2-associated athanogene 6 |
| chr1_+_87620306 | 2.72 |
ENSMUST00000169754.1
|
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr5_-_123879992 | 2.71 |
ENSMUST00000164267.1
|
Gpr81
|
G protein-coupled receptor 81 |
| chr2_+_164805082 | 2.70 |
ENSMUST00000052107.4
|
Zswim3
|
zinc finger SWIM-type containing 3 |
| chr5_+_99979061 | 2.70 |
ENSMUST00000046721.1
|
4930524J08Rik
|
RIKEN cDNA 4930524J08 gene |
| chr8_-_105326252 | 2.68 |
ENSMUST00000070508.7
|
Lrrc29
|
leucine rich repeat containing 29 |
| chr16_-_18811615 | 2.68 |
ENSMUST00000096990.3
|
Cdc45
|
cell division cycle 45 |
| chr16_+_48994185 | 2.66 |
ENSMUST00000117994.1
ENSMUST00000048374.5 |
C330027C09Rik
|
RIKEN cDNA C330027C09 gene |
| chr7_+_126766397 | 2.65 |
ENSMUST00000032944.7
|
Gdpd3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr2_+_22774081 | 2.61 |
ENSMUST00000014290.8
|
Apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr2_+_156840077 | 2.61 |
ENSMUST00000081335.6
ENSMUST00000073352.3 |
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr11_+_61684419 | 2.61 |
ENSMUST00000093019.5
|
Fam83g
|
family with sequence similarity 83, member G |
| chr2_+_71117923 | 2.60 |
ENSMUST00000028403.2
|
Cybrd1
|
cytochrome b reductase 1 |
| chr3_-_129831374 | 2.57 |
ENSMUST00000029643.8
|
Gar1
|
GAR1 ribonucleoprotein homolog (yeast) |
| chr12_-_32208470 | 2.57 |
ENSMUST00000085469.5
|
Pik3cg
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide |
| chr9_+_56418624 | 2.57 |
ENSMUST00000034879.3
|
Hmg20a
|
high mobility group 20A |
| chr17_+_47505117 | 2.55 |
ENSMUST00000183044.1
ENSMUST00000037333.10 |
Ccnd3
|
cyclin D3 |
| chr11_+_101733011 | 2.54 |
ENSMUST00000129741.1
|
Dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr19_-_5986143 | 2.54 |
ENSMUST00000041827.7
|
Slc22a20
|
solute carrier family 22 (organic anion transporter), member 20 |
| chr17_+_47505149 | 2.53 |
ENSMUST00000183177.1
ENSMUST00000182848.1 |
Ccnd3
|
cyclin D3 |
| chr2_+_158768083 | 2.53 |
ENSMUST00000029183.2
|
Fam83d
|
family with sequence similarity 83, member D |
| chr11_+_46454921 | 2.53 |
ENSMUST00000020668.8
|
Havcr2
|
hepatitis A virus cellular receptor 2 |
| chr1_+_135132693 | 2.53 |
ENSMUST00000049449.4
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr2_-_118728345 | 2.52 |
ENSMUST00000159756.1
|
Plcb2
|
phospholipase C, beta 2 |
| chr3_-_105932664 | 2.50 |
ENSMUST00000098758.2
|
I830077J02Rik
|
RIKEN cDNA I830077J02 gene |
| chr4_-_43040279 | 2.45 |
ENSMUST00000107958.1
ENSMUST00000107959.1 ENSMUST00000152846.1 |
Fam214b
|
family with sequence similarity 214, member B |
| chr11_+_46454957 | 2.43 |
ENSMUST00000109229.1
|
Havcr2
|
hepatitis A virus cellular receptor 2 |
| chr17_+_47505211 | 2.41 |
ENSMUST00000182935.1
ENSMUST00000182506.1 |
Ccnd3
|
cyclin D3 |
| chr17_+_35241746 | 2.41 |
ENSMUST00000068056.5
ENSMUST00000174757.1 |
Ddx39b
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chrX_+_48146436 | 2.40 |
ENSMUST00000033427.6
|
Sash3
|
SAM and SH3 domain containing 3 |
| chr5_-_134229581 | 2.39 |
ENSMUST00000111275.1
ENSMUST00000016094.6 ENSMUST00000144086.1 |
Ncf1
|
neutrophil cytosolic factor 1 |
| chr17_+_35135174 | 2.39 |
ENSMUST00000166426.2
ENSMUST00000025250.7 |
Bag6
|
BCL2-associated athanogene 6 |
| chr1_-_173333503 | 2.38 |
ENSMUST00000038227.4
|
Darc
|
Duffy blood group, chemokine receptor |
| chr3_+_105904377 | 2.36 |
ENSMUST00000000574.1
|
Adora3
|
adenosine A3 receptor |
| chr17_-_50094277 | 2.34 |
ENSMUST00000113195.1
|
Rftn1
|
raftlin lipid raft linker 1 |
| chr15_-_98934522 | 2.33 |
ENSMUST00000077577.7
|
Tuba1b
|
tubulin, alpha 1B |
| chr3_-_127409014 | 2.33 |
ENSMUST00000182008.1
ENSMUST00000182711.1 ENSMUST00000182547.1 |
Ank2
|
ankyrin 2, brain |
| chr11_+_3330781 | 2.32 |
ENSMUST00000136536.1
ENSMUST00000093399.4 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr5_+_91517615 | 2.31 |
ENSMUST00000040576.9
|
Parm1
|
prostate androgen-regulated mucin-like protein 1 |
| chr17_+_48462355 | 2.31 |
ENSMUST00000162132.1
|
Unc5cl
|
unc-5 homolog C (C. elegans)-like |
| chr14_-_33185066 | 2.30 |
ENSMUST00000061753.8
ENSMUST00000130509.2 |
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chrX_+_56447965 | 2.30 |
ENSMUST00000079663.6
|
Gm2174
|
predicted gene 2174 |
| chr2_+_84840612 | 2.27 |
ENSMUST00000111625.1
|
Slc43a1
|
solute carrier family 43, member 1 |
| chr11_-_79523760 | 2.24 |
ENSMUST00000179322.1
|
Evi2b
|
ecotropic viral integration site 2b |
| chr17_-_51826562 | 2.23 |
ENSMUST00000024720.4
ENSMUST00000129667.1 ENSMUST00000156051.1 ENSMUST00000169480.1 ENSMUST00000148559.1 |
Satb1
|
special AT-rich sequence binding protein 1 |
| chr3_-_127408986 | 2.23 |
ENSMUST00000182588.1
ENSMUST00000182959.1 ENSMUST00000182452.1 |
Ank2
|
ankyrin 2, brain |
| chr19_-_4191035 | 2.22 |
ENSMUST00000045864.2
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr17_+_7925990 | 2.21 |
ENSMUST00000036370.7
|
Tagap
|
T cell activation Rho GTPase activating protein |
| chr11_+_70130329 | 2.21 |
ENSMUST00000041550.5
ENSMUST00000165951.1 |
Mgl2
|
macrophage galactose N-acetyl-galactosamine specific lectin 2 |
| chr2_-_73485733 | 2.21 |
ENSMUST00000102680.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr9_-_60687459 | 2.20 |
ENSMUST00000114032.1
ENSMUST00000166168.1 ENSMUST00000132366.1 |
Lrrc49
|
leucine rich repeat containing 49 |
| chr9_-_57836706 | 2.20 |
ENSMUST00000164010.1
ENSMUST00000171444.1 ENSMUST00000098686.3 |
Arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr15_+_57985873 | 2.19 |
ENSMUST00000050374.2
|
Fam83a
|
family with sequence similarity 83, member A |
| chr17_+_47594629 | 2.19 |
ENSMUST00000182846.1
|
Ccnd3
|
cyclin D3 |
| chr9_+_70679016 | 2.19 |
ENSMUST00000144537.1
|
Adam10
|
a disintegrin and metallopeptidase domain 10 |
| chr17_+_35135695 | 2.18 |
ENSMUST00000174478.1
ENSMUST00000174281.2 ENSMUST00000173550.1 |
Bag6
|
BCL2-associated athanogene 6 |
| chr3_-_50443603 | 2.18 |
ENSMUST00000029297.4
|
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr7_-_116334132 | 2.17 |
ENSMUST00000170953.1
|
Rps13
|
ribosomal protein S13 |
| chr1_-_133690100 | 2.17 |
ENSMUST00000169295.1
|
Lax1
|
lymphocyte transmembrane adaptor 1 |
| chr2_-_160619971 | 2.17 |
ENSMUST00000109473.1
|
Gm14221
|
predicted gene 14221 |
| chr2_-_91649751 | 2.17 |
ENSMUST00000099714.3
|
Zfp408
|
zinc finger protein 408 |
| chr17_+_35241838 | 2.16 |
ENSMUST00000173731.1
|
Ddx39b
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr9_+_70678950 | 2.16 |
ENSMUST00000067880.6
|
Adam10
|
a disintegrin and metallopeptidase domain 10 |
| chr6_+_125071277 | 2.15 |
ENSMUST00000140346.2
ENSMUST00000171989.1 |
Lpar5
|
lysophosphatidic acid receptor 5 |
| chr7_+_81862674 | 2.15 |
ENSMUST00000119543.1
|
Tm6sf1
|
transmembrane 6 superfamily member 1 |
| chr3_+_131110350 | 2.14 |
ENSMUST00000066849.6
ENSMUST00000106341.2 ENSMUST00000029611.7 |
Lef1
|
lymphoid enhancer binding factor 1 |
| chr4_-_43562397 | 2.13 |
ENSMUST00000030187.7
|
Tln1
|
talin 1 |
| chr5_+_65863563 | 2.12 |
ENSMUST00000031106.4
|
Rhoh
|
ras homolog gene family, member H |
| chr16_-_19983005 | 2.10 |
ENSMUST00000058839.8
|
Klhl6
|
kelch-like 6 |
| chr9_+_107935876 | 2.09 |
ENSMUST00000035700.8
|
Camkv
|
CaM kinase-like vesicle-associated |
| chr15_-_9529868 | 2.09 |
ENSMUST00000003981.4
|
Il7r
|
interleukin 7 receptor |
| chr15_-_66969616 | 2.08 |
ENSMUST00000170903.1
ENSMUST00000166420.1 ENSMUST00000005256.6 ENSMUST00000164070.1 |
Ndrg1
|
N-myc downstream regulated gene 1 |
| chr3_-_127409044 | 2.08 |
ENSMUST00000182704.1
|
Ank2
|
ankyrin 2, brain |
| chr13_+_24614608 | 2.06 |
ENSMUST00000091694.3
|
Fam65b
|
family with sequence similarity 65, member B |
| chr18_+_50030977 | 2.06 |
ENSMUST00000145726.1
ENSMUST00000128377.1 |
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chrX_+_159840463 | 2.05 |
ENSMUST00000112451.1
ENSMUST00000112453.2 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr2_+_29889217 | 2.05 |
ENSMUST00000123335.1
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr14_-_31494992 | 2.05 |
ENSMUST00000055303.3
|
Mettl6
|
methyltransferase like 6 |
| chr2_-_181693810 | 2.04 |
ENSMUST00000108776.1
ENSMUST00000108771.1 ENSMUST00000108779.1 ENSMUST00000108769.1 ENSMUST00000108772.1 ENSMUST00000002532.2 |
Rgs19
|
regulator of G-protein signaling 19 |
| chr6_+_115134899 | 2.04 |
ENSMUST00000009538.5
ENSMUST00000169345.1 |
Syn2
|
synapsin II |
| chr2_+_154200371 | 2.04 |
ENSMUST00000028987.6
|
Bpifb1
|
BPI fold containing family B, member 1 |
| chr6_+_129397478 | 2.03 |
ENSMUST00000112081.2
ENSMUST00000112079.2 |
Clec1b
|
C-type lectin domain family 1, member b |
| chr15_-_66812593 | 2.02 |
ENSMUST00000100572.3
|
Sla
|
src-like adaptor |
| chr9_-_119209096 | 2.02 |
ENSMUST00000084797.4
|
Slc22a13
|
solute carrier family 22 (organic cation transporter), member 13 |
| chr11_+_101732950 | 2.01 |
ENSMUST00000039152.7
|
Dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Etv4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.9 | 35.7 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 3.1 | 9.3 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 2.7 | 8.2 | GO:0030221 | basophil differentiation(GO:0030221) |
| 2.4 | 7.2 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) negative regulation of hypersensitivity(GO:0002884) |
| 2.3 | 9.4 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 2.0 | 8.2 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) meiotic metaphase plate congression(GO:0051311) |
| 1.8 | 8.9 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 1.7 | 6.7 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 1.7 | 5.0 | GO:2001189 | negative regulation of immunological synapse formation(GO:2000521) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 1.4 | 5.4 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 1.3 | 8.0 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 1.3 | 10.4 | GO:0036506 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 1.3 | 3.8 | GO:0015866 | ADP transport(GO:0015866) |
| 1.1 | 8.0 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 1.1 | 7.6 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 1.1 | 3.2 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) negative regulation of extracellular matrix disassembly(GO:0010716) |
| 1.1 | 6.3 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 1.0 | 10.4 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 1.0 | 3.1 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 1.0 | 2.9 | GO:0051659 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.9 | 5.7 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.9 | 11.8 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.9 | 3.6 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.9 | 2.7 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.9 | 6.2 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.9 | 0.9 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.9 | 4.3 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.9 | 7.8 | GO:0036371 | T-tubule organization(GO:0033292) protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.9 | 3.5 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.9 | 11.1 | GO:0046645 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.8 | 4.0 | GO:0071104 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.8 | 7.9 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.8 | 3.1 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.8 | 5.4 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.8 | 2.3 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.8 | 3.8 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.7 | 2.9 | GO:0071608 | macrophage inflammatory protein-1 alpha production(GO:0071608) |
| 0.7 | 2.2 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.7 | 2.8 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.7 | 3.5 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.7 | 4.8 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.7 | 3.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.7 | 4.6 | GO:2000002 | negative regulation of cell cycle checkpoint(GO:1901977) negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.7 | 8.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.6 | 5.8 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.6 | 3.8 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.6 | 4.8 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.5 | 1.6 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.5 | 1.6 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.5 | 9.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.5 | 2.6 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.5 | 1.5 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.5 | 1.0 | GO:1904976 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.5 | 2.0 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.5 | 2.4 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.5 | 0.5 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.5 | 3.7 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.5 | 1.8 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.5 | 1.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.5 | 5.0 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.4 | 1.3 | GO:1904959 | regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.4 | 1.3 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.4 | 1.2 | GO:0043519 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) regulation of bleb assembly(GO:1904170) |
| 0.4 | 0.8 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.4 | 0.8 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.4 | 2.0 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.4 | 1.6 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) |
| 0.4 | 1.2 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.4 | 1.9 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.4 | 5.4 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.4 | 1.1 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.4 | 1.1 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) negative regulation of interleukin-18 production(GO:0032701) |
| 0.4 | 8.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.4 | 1.1 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.4 | 7.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.3 | 1.4 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.3 | 7.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.3 | 7.1 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.3 | 1.3 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.3 | 4.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.3 | 4.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.3 | 2.6 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.3 | 4.6 | GO:0015747 | urate transport(GO:0015747) |
| 0.3 | 0.9 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.3 | 1.6 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.3 | 1.6 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.3 | 1.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.3 | 4.6 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.3 | 3.0 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.3 | 3.8 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.3 | 0.9 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.3 | 0.6 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.3 | 1.4 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.3 | 3.7 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.3 | 2.0 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.3 | 2.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 1.1 | GO:0032962 | regulation of inositol trisphosphate biosynthetic process(GO:0032960) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.3 | 2.0 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.3 | 0.8 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.3 | 1.4 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.3 | 1.1 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.3 | 1.1 | GO:1903527 | regulation of membrane tubulation(GO:1903525) positive regulation of membrane tubulation(GO:1903527) |
| 0.3 | 2.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.3 | 0.5 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.3 | 2.0 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.2 | 6.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 8.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.2 | 1.9 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 1.0 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.2 | 3.4 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.2 | 1.4 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.2 | 1.4 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.2 | 2.1 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.2 | 0.9 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.2 | 3.6 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.2 | 3.2 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.2 | 3.1 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.2 | 1.1 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.2 | 1.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 1.5 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.2 | 2.1 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.2 | 0.6 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.2 | 7.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.2 | 3.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 1.0 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.2 | 7.1 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.2 | 2.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.2 | 1.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 3.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 1.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.2 | 0.6 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.2 | 0.2 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.2 | 0.5 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.2 | 0.5 | GO:2000556 | T-helper 1 cell cytokine production(GO:0035744) regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) |
| 0.2 | 0.5 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.2 | 1.4 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.2 | 19.2 | GO:0030168 | platelet activation(GO:0030168) |
| 0.2 | 1.6 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.2 | 3.5 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.2 | 2.4 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.2 | 1.2 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.2 | 1.9 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 0.2 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.2 | 2.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 1.0 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.2 | 3.0 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.2 | 1.0 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 0.8 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 | 0.6 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) leading strand elongation(GO:0006272) |
| 0.2 | 0.8 | GO:0090290 | regulation of osteoclast proliferation(GO:0090289) positive regulation of osteoclast proliferation(GO:0090290) |
| 0.2 | 1.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 2.6 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.2 | 0.5 | GO:1904579 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.1 | 1.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.3 | GO:0001805 | antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.1 | 1.6 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 2.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.4 | GO:2000314 | negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.1 | 0.6 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 1.4 | GO:0046549 | phenol-containing compound catabolic process(GO:0019336) retinal cone cell development(GO:0046549) |
| 0.1 | 2.1 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.1 | 1.0 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 8.8 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 1.5 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 3.1 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 0.7 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 0.8 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 | 0.7 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.8 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 | 1.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 1.7 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 1.1 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.1 | 1.5 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 1.3 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 0.7 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 1.4 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.1 | 0.9 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 2.0 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 1.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 1.8 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.5 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.1 | 1.4 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 1.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 2.5 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 2.0 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) |
| 0.1 | 0.3 | GO:0071455 | cellular response to hyperoxia(GO:0071455) |
| 0.1 | 3.8 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.1 | 0.4 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.9 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.1 | 0.7 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.1 | 0.5 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 4.0 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.1 | 0.8 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 0.8 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.1 | 0.2 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.1 | 0.5 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.5 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 0.5 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.1 | 2.7 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.1 | 0.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.7 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.1 | 3.2 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 1.6 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 0.4 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.4 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.1 | 0.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 1.8 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 0.3 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 0.5 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 0.6 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 3.4 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.1 | 0.2 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.1 | 0.7 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 1.8 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.1 | 1.2 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 0.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 3.0 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.1 | 1.2 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 1.0 | GO:0051444 | negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.1 | 2.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 1.7 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 2.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 1.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.7 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.6 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 0.3 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.5 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.3 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.8 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.1 | 2.6 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 0.7 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.1 | 2.1 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.1 | 1.1 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.1 | 0.3 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 2.3 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.9 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.1 | 0.3 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 1.4 | GO:0071353 | response to interleukin-4(GO:0070670) cellular response to interleukin-4(GO:0071353) |
| 0.1 | 0.5 | GO:0033127 | regulation of histone phosphorylation(GO:0033127) |
| 0.1 | 3.1 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 2.7 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.0 | 1.5 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.3 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.8 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 1.5 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 1.9 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.9 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.6 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.4 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.4 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.6 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.1 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.1 | GO:2000584 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.4 | GO:0036260 | 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.9 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 1.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 2.8 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.5 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.5 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 1.4 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 | 0.3 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.3 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 3.9 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.8 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.7 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 1.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.5 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.4 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.0 | 0.2 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.6 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.5 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.7 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.2 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.2 | GO:0032099 | negative regulation of appetite(GO:0032099) |
| 0.0 | 0.4 | GO:0048753 | melanosome organization(GO:0032438) pigment granule organization(GO:0048753) |
| 0.0 | 1.2 | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 | 0.3 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.0 | GO:2000078 | columnar/cuboidal epithelial cell maturation(GO:0002069) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 0.2 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 33.5 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 1.6 | 6.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 1.5 | 10.4 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 1.3 | 7.8 | GO:0097443 | sorting endosome(GO:0097443) |
| 1.3 | 6.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 1.1 | 6.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 1.0 | 6.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.9 | 2.8 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.9 | 2.7 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.7 | 9.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.7 | 2.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.7 | 8.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.7 | 8.0 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.6 | 5.7 | GO:0042581 | specific granule(GO:0042581) |
| 0.6 | 7.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.6 | 10.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.6 | 9.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.6 | 4.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.5 | 3.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.5 | 1.5 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.5 | 5.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.5 | 3.0 | GO:0031673 | H zone(GO:0031673) |
| 0.5 | 2.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.5 | 2.4 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.5 | 3.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.4 | 2.6 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.4 | 1.7 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.4 | 0.8 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.4 | 3.1 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.4 | 3.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 1.4 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.3 | 0.9 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.3 | 0.9 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.3 | 7.1 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.3 | 0.9 | GO:0036019 | endolysosome(GO:0036019) |
| 0.3 | 2.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.3 | 1.6 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.3 | 6.7 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.3 | 1.3 | GO:1990745 | EARP complex(GO:1990745) |
| 0.3 | 11.1 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 3.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.2 | 2.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 1.9 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 9.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.2 | 13.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 4.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 9.4 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 1.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 2.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 1.9 | GO:0043219 | lateral loop(GO:0043219) |
| 0.2 | 0.8 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.2 | 14.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 2.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.2 | 8.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 2.4 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.2 | 1.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 0.9 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 3.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 0.9 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 6.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 2.0 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 2.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 0.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 2.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 2.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 2.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 1.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.6 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 1.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 7.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 3.2 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 1.8 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 3.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 7.1 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 1.8 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 1.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 2.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 13.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 0.5 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.1 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.7 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 7.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 2.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 3.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 3.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 7.5 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 1.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 2.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.2 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.1 | 4.0 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 0.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 0.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 2.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 1.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.0 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 2.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 5.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 2.4 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 13.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.1 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 2.8 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 1.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.8 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 2.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 7.9 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.5 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.3 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 1.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 1.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.0 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.2 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 2.3 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 1.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 3.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 2.0 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0071565 | nBAF complex(GO:0071565) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.2 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 2.7 | 8.0 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) |
| 2.6 | 7.7 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 2.6 | 10.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.8 | 7.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 1.7 | 8.4 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 1.4 | 5.7 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 1.3 | 6.5 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 1.0 | 9.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 1.0 | 9.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 1.0 | 2.9 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 1.0 | 4.8 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.9 | 5.4 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.9 | 6.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.8 | 9.3 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.8 | 0.8 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.7 | 2.8 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.7 | 15.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.7 | 5.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.6 | 1.9 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.6 | 2.5 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.6 | 1.8 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.6 | 2.3 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.6 | 6.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.6 | 35.5 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.5 | 8.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.5 | 4.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.5 | 8.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.5 | 1.4 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.5 | 1.4 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.4 | 1.3 | GO:0070138 | isopeptidase activity(GO:0070122) ubiquitin-like protein-specific isopeptidase activity(GO:0070138) SUMO-specific isopeptidase activity(GO:0070140) |
| 0.4 | 1.3 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.4 | 1.2 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.4 | 1.6 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) prostaglandin-E synthase activity(GO:0050220) |
| 0.4 | 1.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.4 | 2.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.4 | 2.2 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.4 | 4.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.4 | 2.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.3 | 2.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.3 | 2.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.3 | 3.8 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 7.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.3 | 1.8 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.3 | 10.0 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.3 | 1.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 0.9 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.3 | 3.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.3 | 4.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.3 | 4.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.3 | 1.3 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.3 | 5.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 1.7 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.2 | 1.0 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.2 | 2.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.2 | 1.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.2 | 2.9 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.2 | 1.7 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.2 | 1.9 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.2 | 0.9 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.2 | 0.7 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 2.9 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 0.4 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.2 | 3.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 7.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 0.9 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.2 | 2.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 0.8 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.2 | 3.2 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.2 | 10.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 5.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 2.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 3.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 0.5 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.2 | 2.8 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.2 | 0.5 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.2 | 0.9 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 0.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 4.1 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.2 | 7.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 3.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 5.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 2.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 7.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 1.0 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 3.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 1.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 0.5 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 2.0 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 6.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.4 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 2.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.1 | 9.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 4.6 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 3.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 3.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.5 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.9 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 2.6 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 2.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.7 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.1 | 4.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.6 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 1.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 1.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 1.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 1.7 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.8 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 3.0 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 6.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.8 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.3 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 2.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 6.5 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.3 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 1.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 13.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.6 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 0.9 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 1.0 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.3 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 1.8 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.1 | 3.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 2.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 1.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 1.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.3 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 0.3 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.6 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.1 | 0.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 2.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 7.1 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 2.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 8.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 2.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.9 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 2.0 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.7 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 2.8 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.5 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.9 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 3.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 11.4 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.8 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.3 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 1.9 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 1.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 4.4 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 7.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 19.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.8 | 42.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.6 | 7.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.4 | 15.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.4 | 6.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.4 | 2.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.2 | 7.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 15.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.2 | 11.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 9.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 8.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 10.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 5.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 3.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 7.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 2.8 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.1 | 6.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 10.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 6.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 8.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 4.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 4.0 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 3.6 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 10.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 2.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 7.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 1.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 3.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.2 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 0.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 1.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 1.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 2.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 2.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 11.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 0.9 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.5 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 2.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 2.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 6.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 15.9 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.6 | 6.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.6 | 8.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.6 | 6.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.4 | 5.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 5.0 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.4 | 4.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.4 | 5.9 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.4 | 2.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.3 | 1.0 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.3 | 4.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.3 | 9.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.3 | 13.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.2 | 25.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 9.9 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.2 | 6.9 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 3.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 2.9 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.2 | 7.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.2 | 2.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 5.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.2 | 1.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 2.6 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 3.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 2.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 3.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 2.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 2.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 13.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 4.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 11.4 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.1 | 2.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 3.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 2.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 4.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 5.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 0.8 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 0.9 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.0 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 1.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.8 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 7.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 2.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 4.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 3.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.9 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 7.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 2.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.3 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 4.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.6 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.0 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.9 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 2.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.9 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |