Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Foxl1
Z-value: 0.57
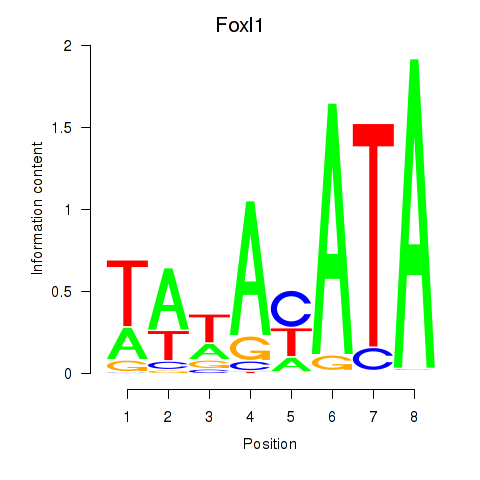
Transcription factors associated with Foxl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxl1
|
ENSMUSG00000097084.1 | forkhead box L1 |
|
Foxl1
|
ENSMUSG00000043867.5 | forkhead box L1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxl1 | mm10_v2_chr8_+_121127827_121127940 | 0.59 | 1.5e-04 | Click! |
Activity profile of Foxl1 motif
Sorted Z-values of Foxl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_40811089 | 5.07 |
ENSMUST00000024721.7
|
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr4_-_99654983 | 4.19 |
ENSMUST00000136525.1
|
Gm12688
|
predicted gene 12688 |
| chr10_+_79879614 | 4.02 |
ENSMUST00000006679.8
|
Prtn3
|
proteinase 3 |
| chr13_+_23763660 | 2.83 |
ENSMUST00000055770.1
|
Hist1h1a
|
histone cluster 1, H1a |
| chr4_+_11191726 | 2.60 |
ENSMUST00000029866.9
ENSMUST00000108324.3 |
Ccne2
|
cyclin E2 |
| chr3_-_123034943 | 2.50 |
ENSMUST00000029761.7
|
Myoz2
|
myozenin 2 |
| chr8_+_15057646 | 2.28 |
ENSMUST00000033842.3
|
Myom2
|
myomesin 2 |
| chr13_+_23575753 | 2.07 |
ENSMUST00000105105.1
|
Hist1h3d
|
histone cluster 1, H3d |
| chr10_+_88091070 | 2.05 |
ENSMUST00000048621.7
|
Pmch
|
pro-melanin-concentrating hormone |
| chr9_+_121777607 | 1.48 |
ENSMUST00000098272.2
|
Klhl40
|
kelch-like 40 |
| chr18_-_15151427 | 1.48 |
ENSMUST00000025992.6
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chr11_+_31872100 | 1.43 |
ENSMUST00000020543.6
ENSMUST00000109412.2 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr9_+_71215779 | 1.21 |
ENSMUST00000034723.5
|
Aldh1a2
|
aldehyde dehydrogenase family 1, subfamily A2 |
| chr13_-_23622502 | 1.14 |
ENSMUST00000062045.2
|
Hist1h1e
|
histone cluster 1, H1e |
| chr7_+_91090728 | 1.06 |
ENSMUST00000074273.3
|
Dlg2
|
discs, large homolog 2 (Drosophila) |
| chrX_+_133850980 | 1.06 |
ENSMUST00000033602.8
|
Tnmd
|
tenomodulin |
| chr7_+_91090697 | 1.01 |
ENSMUST00000107196.2
|
Dlg2
|
discs, large homolog 2 (Drosophila) |
| chr12_+_69963452 | 0.97 |
ENSMUST00000110560.1
|
Gm3086
|
predicted gene 3086 |
| chr2_-_59948155 | 0.94 |
ENSMUST00000153136.1
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr13_+_23555023 | 0.83 |
ENSMUST00000045301.6
|
Hist1h1d
|
histone cluster 1, H1d |
| chrX_+_112604274 | 0.81 |
ENSMUST00000071814.6
|
Zfp711
|
zinc finger protein 711 |
| chr18_+_34409415 | 0.79 |
ENSMUST00000166156.1
ENSMUST00000014647.7 |
Pkd2l2
|
polycystic kidney disease 2-like 2 |
| chr13_+_23738804 | 0.72 |
ENSMUST00000040914.1
|
Hist1h1c
|
histone cluster 1, H1c |
| chr7_-_119479249 | 0.70 |
ENSMUST00000033263.4
|
Umod
|
uromodulin |
| chr1_+_66386968 | 0.68 |
ENSMUST00000145419.1
|
Map2
|
microtubule-associated protein 2 |
| chr5_-_83355120 | 0.63 |
ENSMUST00000053543.4
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr7_+_122289297 | 0.63 |
ENSMUST00000064989.5
ENSMUST00000064921.4 |
Prkcb
|
protein kinase C, beta |
| chr6_-_142804390 | 0.58 |
ENSMUST00000111768.1
|
Gm766
|
predicted gene 766 |
| chr4_-_121017201 | 0.51 |
ENSMUST00000043200.7
|
Smap2
|
small ArfGAP 2 |
| chr5_+_33018816 | 0.37 |
ENSMUST00000019109.7
|
Ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
| chr6_+_42286676 | 0.28 |
ENSMUST00000031894.6
|
Clcn1
|
chloride channel 1 |
| chr15_+_43477213 | 0.26 |
ENSMUST00000022962.6
|
Emc2
|
ER membrane protein complex subunit 2 |
| chrX_-_94123087 | 0.25 |
ENSMUST00000113925.1
|
Zfx
|
zinc finger protein X-linked |
| chr7_+_84853573 | 0.25 |
ENSMUST00000078172.4
|
Olfr291
|
olfactory receptor 291 |
| chr5_-_83354946 | 0.25 |
ENSMUST00000146669.1
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr15_-_11905609 | 0.20 |
ENSMUST00000066529.3
|
Npr3
|
natriuretic peptide receptor 3 |
| chrX_+_18162575 | 0.15 |
ENSMUST00000044484.6
ENSMUST00000052368.8 |
Kdm6a
|
lysine (K)-specific demethylase 6A |
| chr2_+_18055203 | 0.14 |
ENSMUST00000028076.8
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr6_+_42286709 | 0.14 |
ENSMUST00000163936.1
|
Clcn1
|
chloride channel 1 |
| chr10_+_56377300 | 0.08 |
ENSMUST00000068581.7
|
Gja1
|
gap junction protein, alpha 1 |
| chr5_-_87699414 | 0.06 |
ENSMUST00000082370.5
|
Csn2
|
casein beta |
| chr1_-_144249134 | 0.05 |
ENSMUST00000172388.1
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr6_-_14755250 | 0.03 |
ENSMUST00000045096.4
|
Ppp1r3a
|
protein phosphatase 1, regulatory (inhibitor) subunit 3A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.8 | 4.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.7 | 2.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.3 | 2.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.3 | 1.5 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.2 | 0.7 | GO:0072236 | metanephric loop of Henle development(GO:0072236) |
| 0.2 | 1.2 | GO:0042905 | ureter maturation(GO:0035799) 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 2.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 1.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 2.1 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.1 | 0.6 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.4 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.1 | 2.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 2.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.9 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.4 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 1.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 2.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 2.5 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.1 | GO:0010643 | positive regulation of glomerular filtration(GO:0003104) cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.2 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.7 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 2.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 7.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 2.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.7 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 1.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.5 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 2.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.3 | 5.1 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.2 | 1.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 1.1 | GO:0032564 | dATP binding(GO:0032564) |
| 0.1 | 2.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.6 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.7 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 2.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.7 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 1.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 2.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 2.1 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 2.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 1.6 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 2.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 3.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.0 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 5.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 2.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 2.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.6 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |