Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Gata2_Gata1
Z-value: 4.26
Transcription factors associated with Gata2_Gata1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
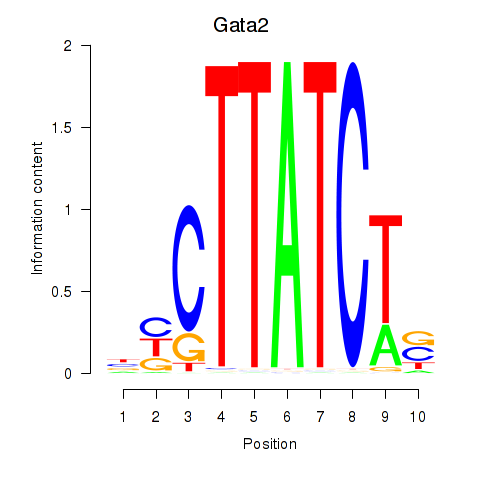
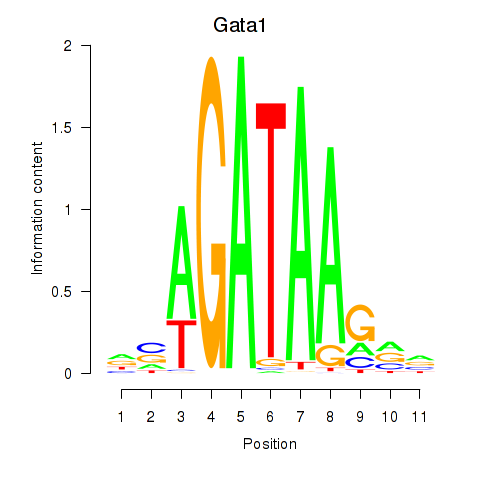
Gata2
|
ENSMUSG00000015053.8 | GATA binding protein 2 |
|
Gata1
|
ENSMUSG00000031162.8 | GATA binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gata1 | mm10_v2_chrX_-_7964166_7964191 | 0.89 | 2.4e-13 | Click! |
| Gata2 | mm10_v2_chr6_+_88198656_88198675 | 0.83 | 3.6e-10 | Click! |
Activity profile of Gata2_Gata1 motif
Sorted Z-values of Gata2_Gata1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_26199008 | 69.41 |
ENSMUST00000142410.1
ENSMUST00000120333.1 ENSMUST00000039113.7 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr4_-_46404224 | 64.49 |
ENSMUST00000107764.2
|
Hemgn
|
hemogen |
| chr6_+_41458923 | 59.97 |
ENSMUST00000031910.7
|
Prss1
|
protease, serine, 1 (trypsin 1) |
| chr2_+_84980458 | 57.88 |
ENSMUST00000028467.5
|
Prg2
|
proteoglycan 2, bone marrow |
| chr2_-_121036750 | 55.99 |
ENSMUST00000023987.5
|
Epb4.2
|
erythrocyte protein band 4.2 |
| chr11_+_32276400 | 54.81 |
ENSMUST00000020531.2
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr6_+_30639218 | 54.60 |
ENSMUST00000031806.9
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr6_-_41704339 | 52.84 |
ENSMUST00000031899.8
|
Kel
|
Kell blood group |
| chr6_+_41392356 | 52.67 |
ENSMUST00000049079.7
|
Gm5771
|
predicted gene 5771 |
| chr8_+_84701430 | 51.74 |
ENSMUST00000037165.4
|
Lyl1
|
lymphoblastomic leukemia 1 |
| chr2_-_121037048 | 50.08 |
ENSMUST00000102490.3
|
Epb4.2
|
erythrocyte protein band 4.2 |
| chr17_-_35085609 | 47.50 |
ENSMUST00000038507.6
|
Ly6g6f
|
lymphocyte antigen 6 complex, locus G6F |
| chr17_+_36869567 | 47.36 |
ENSMUST00000060524.9
|
Trim10
|
tripartite motif-containing 10 |
| chr3_-_84479418 | 45.16 |
ENSMUST00000091002.1
|
Fhdc1
|
FH2 domain containing 1 |
| chrX_-_7964166 | 45.09 |
ENSMUST00000128449.1
|
Gata1
|
GATA binding protein 1 |
| chr5_-_73191848 | 44.94 |
ENSMUST00000176910.1
|
Fryl
|
furry homolog-like (Drosophila) |
| chr1_-_132367879 | 44.73 |
ENSMUST00000142609.1
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr11_+_32276893 | 43.45 |
ENSMUST00000145569.1
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr17_-_28560704 | 42.20 |
ENSMUST00000114785.1
ENSMUST00000025062.3 |
Clps
|
colipase, pancreatic |
| chr8_-_85380964 | 41.40 |
ENSMUST00000122452.1
|
Mylk3
|
myosin light chain kinase 3 |
| chr9_+_21029373 | 41.09 |
ENSMUST00000001040.5
|
Icam4
|
intercellular adhesion molecule 4, Landsteiner-Wiener blood group |
| chr6_+_30541582 | 40.98 |
ENSMUST00000096066.4
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr14_+_27000362 | 38.94 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr11_-_83286722 | 38.86 |
ENSMUST00000163961.2
|
Slfn14
|
schlafen family member 14 |
| chr17_-_35066170 | 38.73 |
ENSMUST00000174190.1
ENSMUST00000097337.1 |
AU023871
|
expressed sequence AU023871 |
| chr4_+_134864536 | 38.46 |
ENSMUST00000030627.7
|
Rhd
|
Rh blood group, D antigen |
| chr18_-_78123324 | 37.98 |
ENSMUST00000160292.1
ENSMUST00000091813.5 |
Slc14a1
|
solute carrier family 14 (urea transporter), member 1 |
| chr4_-_137430517 | 37.43 |
ENSMUST00000102522.4
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr6_+_86078070 | 37.28 |
ENSMUST00000032069.5
|
Add2
|
adducin 2 (beta) |
| chr3_-_20242173 | 37.10 |
ENSMUST00000001921.1
|
Cpa3
|
carboxypeptidase A3, mast cell |
| chr15_-_103251465 | 33.67 |
ENSMUST00000133600.1
ENSMUST00000134554.1 ENSMUST00000156927.1 ENSMUST00000149111.1 ENSMUST00000132836.1 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr10_-_62342674 | 33.44 |
ENSMUST00000143179.1
ENSMUST00000130422.1 |
Hk1
|
hexokinase 1 |
| chr8_+_84901928 | 32.25 |
ENSMUST00000067060.7
|
Klf1
|
Kruppel-like factor 1 (erythroid) |
| chr7_-_4397705 | 32.10 |
ENSMUST00000108590.2
|
Gp6
|
glycoprotein 6 (platelet) |
| chr9_-_21963568 | 31.31 |
ENSMUST00000006397.5
|
Epor
|
erythropoietin receptor |
| chr6_+_41302265 | 30.91 |
ENSMUST00000031913.4
|
Try4
|
trypsin 4 |
| chr10_-_80421847 | 30.43 |
ENSMUST00000156244.1
|
Tcf3
|
transcription factor 3 |
| chr7_-_99238564 | 28.39 |
ENSMUST00000064231.7
|
Mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr5_+_115466234 | 28.03 |
ENSMUST00000145785.1
ENSMUST00000031495.4 ENSMUST00000112071.1 ENSMUST00000125568.1 |
Pla2g1b
|
phospholipase A2, group IB, pancreas |
| chr2_+_103957976 | 26.84 |
ENSMUST00000156813.1
ENSMUST00000170926.1 |
Lmo2
|
LIM domain only 2 |
| chr7_-_100467149 | 26.50 |
ENSMUST00000184420.1
|
RP23-308M1.2
|
RP23-308M1.2 |
| chr16_+_38362205 | 25.31 |
ENSMUST00000023494.6
|
Popdc2
|
popeye domain containing 2 |
| chrX_+_93675088 | 24.02 |
ENSMUST00000045898.3
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr11_+_95337012 | 23.53 |
ENSMUST00000037502.6
|
Fam117a
|
family with sequence similarity 117, member A |
| chr11_-_87359011 | 23.47 |
ENSMUST00000055438.4
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing) |
| chr6_-_41314700 | 23.33 |
ENSMUST00000064324.5
|
Try5
|
trypsin 5 |
| chr16_+_38362234 | 22.45 |
ENSMUST00000114739.1
|
Popdc2
|
popeye domain containing 2 |
| chr4_-_137409777 | 22.41 |
ENSMUST00000024200.6
|
Gm13011
|
predicted gene 13011 |
| chr1_+_40429563 | 22.24 |
ENSMUST00000174335.1
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr11_-_116076986 | 22.23 |
ENSMUST00000153408.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr11_-_102469839 | 22.03 |
ENSMUST00000103086.3
|
Itga2b
|
integrin alpha 2b |
| chr17_-_25944932 | 21.58 |
ENSMUST00000085027.3
|
Nhlrc4
|
NHL repeat containing 4 |
| chr4_+_119637704 | 21.06 |
ENSMUST00000024015.2
|
Guca2a
|
guanylate cyclase activator 2a (guanylin) |
| chr6_-_122609964 | 20.95 |
ENSMUST00000032211.4
|
Gdf3
|
growth differentiation factor 3 |
| chr11_-_99438143 | 19.95 |
ENSMUST00000017743.2
|
Krt20
|
keratin 20 |
| chrX_-_107403295 | 19.76 |
ENSMUST00000033591.5
|
Itm2a
|
integral membrane protein 2A |
| chr4_-_119189949 | 19.53 |
ENSMUST00000124626.1
|
Ermap
|
erythroblast membrane-associated protein |
| chr7_+_28540863 | 19.22 |
ENSMUST00000119180.2
|
Sycn
|
syncollin |
| chr8_-_85365341 | 19.00 |
ENSMUST00000121972.1
|
Mylk3
|
myosin light chain kinase 3 |
| chr8_-_85365317 | 18.99 |
ENSMUST00000034133.7
|
Mylk3
|
myosin light chain kinase 3 |
| chr19_-_7019423 | 18.98 |
ENSMUST00000040772.8
|
Fermt3
|
fermitin family homolog 3 (Drosophila) |
| chr4_+_13751297 | 18.50 |
ENSMUST00000105566.2
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr2_-_153241402 | 18.10 |
ENSMUST00000056924.7
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr3_+_108364882 | 16.04 |
ENSMUST00000090563.5
|
Mybphl
|
myosin binding protein H-like |
| chr4_+_40920047 | 15.96 |
ENSMUST00000030122.4
|
Spink4
|
serine peptidase inhibitor, Kazal type 4 |
| chr9_-_44342332 | 14.95 |
ENSMUST00000097558.3
|
Hmbs
|
hydroxymethylbilane synthase |
| chr13_-_55528511 | 14.72 |
ENSMUST00000047877.4
|
Dok3
|
docking protein 3 |
| chr7_+_110773658 | 13.87 |
ENSMUST00000143786.1
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr15_+_80623499 | 13.75 |
ENSMUST00000043149.7
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr15_+_57985873 | 13.34 |
ENSMUST00000050374.2
|
Fam83a
|
family with sequence similarity 83, member A |
| chr19_+_10015016 | 13.19 |
ENSMUST00000137637.1
ENSMUST00000149967.1 |
Rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr3_-_107221722 | 13.14 |
ENSMUST00000029504.8
|
Cym
|
chymosin |
| chr4_-_119190005 | 13.03 |
ENSMUST00000138395.1
ENSMUST00000156746.1 |
Ermap
|
erythroblast membrane-associated protein |
| chr14_+_75455957 | 12.70 |
ENSMUST00000164848.1
|
Siah3
|
seven in absentia homolog 3 (Drosophila) |
| chr11_-_116077954 | 11.77 |
ENSMUST00000106451.1
ENSMUST00000075036.2 |
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr1_-_173333503 | 11.53 |
ENSMUST00000038227.4
|
Darc
|
Duffy blood group, chemokine receptor |
| chr11_-_116077562 | 11.30 |
ENSMUST00000174822.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr2_+_72476225 | 11.07 |
ENSMUST00000157019.1
|
Cdca7
|
cell division cycle associated 7 |
| chrX_+_150547375 | 11.07 |
ENSMUST00000066337.6
ENSMUST00000112715.1 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr11_-_116077927 | 11.03 |
ENSMUST00000156545.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr2_+_72476159 | 11.01 |
ENSMUST00000102691.4
|
Cdca7
|
cell division cycle associated 7 |
| chr10_-_62327757 | 10.97 |
ENSMUST00000139228.1
|
Hk1
|
hexokinase 1 |
| chr11_-_116077606 | 10.53 |
ENSMUST00000106450.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr7_-_119459266 | 10.48 |
ENSMUST00000033255.5
|
Gp2
|
glycoprotein 2 (zymogen granule membrane) |
| chr11_+_70639118 | 10.43 |
ENSMUST00000055184.6
ENSMUST00000108551.2 |
Gp1ba
|
glycoprotein 1b, alpha polypeptide |
| chr19_-_10678001 | 10.33 |
ENSMUST00000025647.5
|
Pga5
|
pepsinogen 5, group I |
| chr11_+_70505244 | 10.31 |
ENSMUST00000019063.2
|
Tm4sf5
|
transmembrane 4 superfamily member 5 |
| chr1_+_75507077 | 9.33 |
ENSMUST00000037330.4
|
Inha
|
inhibin alpha |
| chr17_-_33713372 | 9.06 |
ENSMUST00000173392.1
|
March2
|
membrane-associated ring finger (C3HC4) 2 |
| chr13_-_73700721 | 9.04 |
ENSMUST00000022048.5
|
Slc6a19
|
solute carrier family 6 (neurotransmitter transporter), member 19 |
| chr6_-_136922169 | 8.63 |
ENSMUST00000032343.6
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr3_-_75270073 | 8.63 |
ENSMUST00000039047.4
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr6_+_87428986 | 8.30 |
ENSMUST00000032125.5
|
Bmp10
|
bone morphogenetic protein 10 |
| chr7_+_28833975 | 8.01 |
ENSMUST00000066723.8
|
Lgals4
|
lectin, galactose binding, soluble 4 |
| chr3_+_79884576 | 7.97 |
ENSMUST00000145992.1
|
Fam198b
|
family with sequence similarity 198, member B |
| chr11_+_87760533 | 7.48 |
ENSMUST00000039627.5
ENSMUST00000100644.3 |
Bzrap1
|
benzodiazepine receptor associated protein 1 |
| chr10_+_80019621 | 7.37 |
ENSMUST00000043311.6
|
Hmha1
|
histocompatibility (minor) HA-1 |
| chr11_-_94242701 | 7.10 |
ENSMUST00000061469.3
|
Wfikkn2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr16_+_44943678 | 6.97 |
ENSMUST00000114613.2
ENSMUST00000114612.1 ENSMUST00000077178.6 ENSMUST00000048479.7 ENSMUST00000114611.3 ENSMUST00000164007.1 ENSMUST00000171779.1 |
Cd200r3
|
CD200 receptor 3 |
| chr11_+_4986824 | 6.84 |
ENSMUST00000009234.9
ENSMUST00000109897.1 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr1_-_138175238 | 6.82 |
ENSMUST00000182536.1
|
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr6_+_125552948 | 6.61 |
ENSMUST00000112254.1
ENSMUST00000112253.1 ENSMUST00000001995.7 |
Vwf
|
Von Willebrand factor homolog |
| chr8_+_120488416 | 6.46 |
ENSMUST00000034279.9
|
Gse1
|
genetic suppressor element 1 |
| chr7_-_141655319 | 6.45 |
ENSMUST00000062451.7
|
Muc6
|
mucin 6, gastric |
| chr6_-_136941887 | 6.41 |
ENSMUST00000111891.1
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr1_-_138175283 | 6.23 |
ENSMUST00000182755.1
ENSMUST00000183262.1 ENSMUST00000027645.7 ENSMUST00000112036.2 ENSMUST00000182283.1 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr1_-_138175126 | 6.09 |
ENSMUST00000183301.1
|
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr3_+_105904377 | 5.73 |
ENSMUST00000000574.1
|
Adora3
|
adenosine A3 receptor |
| chr9_+_66350465 | 5.50 |
ENSMUST00000042824.6
|
Herc1
|
hect (homologous to the E6-AP (UBE3A) carboxyl terminus) domain and RCC1 (CHC1)-like domain (RLD) 1 |
| chr2_+_164403194 | 5.09 |
ENSMUST00000017151.1
|
Rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr4_+_150148905 | 5.08 |
ENSMUST00000059893.7
|
Slc2a7
|
solute carrier family 2 (facilitated glucose transporter), member 7 |
| chr6_-_136941494 | 4.97 |
ENSMUST00000111892.1
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr15_+_76904070 | 4.92 |
ENSMUST00000004072.8
|
Rpl8
|
ribosomal protein L8 |
| chr6_-_136941694 | 4.82 |
ENSMUST00000032344.5
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr9_-_32541589 | 4.54 |
ENSMUST00000016231.7
|
Fli1
|
Friend leukemia integration 1 |
| chr19_-_6015152 | 4.23 |
ENSMUST00000025891.8
|
Capn1
|
calpain 1 |
| chr17_+_46650328 | 4.16 |
ENSMUST00000043464.7
|
Cul7
|
cullin 7 |
| chr11_-_99322943 | 3.89 |
ENSMUST00000038004.2
|
Krt25
|
keratin 25 |
| chr8_+_120537423 | 3.72 |
ENSMUST00000118136.1
|
Gse1
|
genetic suppressor element 1 |
| chr3_+_79884496 | 3.69 |
ENSMUST00000118853.1
|
Fam198b
|
family with sequence similarity 198, member B |
| chr15_-_98607611 | 3.67 |
ENSMUST00000096224.4
|
Adcy6
|
adenylate cyclase 6 |
| chr18_+_76059458 | 3.66 |
ENSMUST00000167921.1
|
Zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr10_+_69785507 | 3.52 |
ENSMUST00000182993.1
|
Ank3
|
ankyrin 3, epithelial |
| chrX_-_73869804 | 3.43 |
ENSMUST00000066576.5
ENSMUST00000114430.1 |
L1cam
|
L1 cell adhesion molecule |
| chr19_-_6015769 | 3.43 |
ENSMUST00000164843.1
|
Capn1
|
calpain 1 |
| chr2_-_33087169 | 3.37 |
ENSMUST00000102810.3
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr3_-_14808358 | 3.37 |
ENSMUST00000181860.1
ENSMUST00000144327.2 |
Car1
|
carbonic anhydrase 1 |
| chr17_+_32621319 | 3.35 |
ENSMUST00000077639.5
|
Gm9705
|
predicted gene 9705 |
| chr3_-_63851251 | 3.32 |
ENSMUST00000162269.2
ENSMUST00000159676.2 ENSMUST00000175947.1 |
Plch1
|
phospholipase C, eta 1 |
| chr11_-_26591729 | 3.21 |
ENSMUST00000109504.1
|
Vrk2
|
vaccinia related kinase 2 |
| chr2_+_131133497 | 3.09 |
ENSMUST00000110225.1
|
Gm11037
|
predicted gene 11037 |
| chr19_-_10482874 | 3.07 |
ENSMUST00000038842.3
|
Ppp1r32
|
protein phosphatase 1, regulatory subunit 32 |
| chr11_+_90030295 | 2.93 |
ENSMUST00000092788.3
|
Tmem100
|
transmembrane protein 100 |
| chr17_-_31144271 | 2.85 |
ENSMUST00000024826.7
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr11_-_86357570 | 2.84 |
ENSMUST00000043624.8
|
Med13
|
mediator complex subunit 13 |
| chr16_-_63864114 | 2.83 |
ENSMUST00000064405.6
|
Epha3
|
Eph receptor A3 |
| chr4_-_32923455 | 2.34 |
ENSMUST00000035719.4
ENSMUST00000084749.1 |
Ankrd6
|
ankyrin repeat domain 6 |
| chr7_-_98112618 | 2.26 |
ENSMUST00000153657.1
|
Myo7a
|
myosin VIIA |
| chr7_-_135528645 | 2.12 |
ENSMUST00000053716.7
|
Clrn3
|
clarin 3 |
| chr3_-_144819494 | 2.07 |
ENSMUST00000029929.7
|
Clca2
|
chloride channel calcium activated 2 |
| chr14_+_66784523 | 2.05 |
ENSMUST00000071522.2
|
Gm10032
|
predicted gene 10032 |
| chr1_+_191025350 | 2.03 |
ENSMUST00000181050.1
|
A230020J21Rik
|
RIKEN cDNA A230020J21 gene |
| chr6_+_129397478 | 1.95 |
ENSMUST00000112081.2
ENSMUST00000112079.2 |
Clec1b
|
C-type lectin domain family 1, member b |
| chr3_-_94473591 | 1.95 |
ENSMUST00000029785.3
|
Riiad1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 |
| chr10_+_116177351 | 1.87 |
ENSMUST00000155606.1
ENSMUST00000128399.1 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr2_-_33086366 | 1.86 |
ENSMUST00000049618.2
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chrX_-_7572843 | 1.79 |
ENSMUST00000132788.1
|
Ppp1r3f
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F |
| chr3_+_138352378 | 1.72 |
ENSMUST00000090166.4
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr3_+_96596628 | 1.68 |
ENSMUST00000058943.7
|
Ankrd34a
|
ankyrin repeat domain 34A |
| chrX_+_73757069 | 1.63 |
ENSMUST00000002079.6
|
Plxnb3
|
plexin B3 |
| chr14_-_54653616 | 1.63 |
ENSMUST00000126166.1
ENSMUST00000141453.1 ENSMUST00000150371.1 ENSMUST00000123875.1 ENSMUST00000022794.7 ENSMUST00000148754.3 |
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr11_-_53707269 | 1.63 |
ENSMUST00000124352.1
ENSMUST00000020649.7 |
Rad50
|
RAD50 homolog (S. cerevisiae) |
| chrX_+_166238901 | 1.62 |
ENSMUST00000112235.1
|
Gpm6b
|
glycoprotein m6b |
| chr16_+_25801907 | 1.62 |
ENSMUST00000040231.6
ENSMUST00000115306.1 ENSMUST00000115304.1 ENSMUST00000115305.1 |
Trp63
|
transformation related protein 63 |
| chr2_+_32525013 | 1.60 |
ENSMUST00000150621.1
|
Gm13412
|
predicted gene 13412 |
| chr11_+_96024540 | 1.57 |
ENSMUST00000103157.3
|
Gip
|
gastric inhibitory polypeptide |
| chr12_-_69893162 | 1.56 |
ENSMUST00000049239.7
ENSMUST00000110570.1 |
Map4k5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr3_-_102782708 | 1.53 |
ENSMUST00000029450.3
ENSMUST00000172026.1 ENSMUST00000170856.1 |
Tshb
|
thyroid stimulating hormone, beta subunit |
| chr2_-_25224653 | 1.40 |
ENSMUST00000043584.4
|
Tubb4b
|
tubulin, beta 4B class IVB |
| chr5_+_24364804 | 1.37 |
ENSMUST00000030834.4
ENSMUST00000115090.1 |
Nos3
|
nitric oxide synthase 3, endothelial cell |
| chr6_-_71632897 | 1.27 |
ENSMUST00000065509.4
|
Kdm3a
|
lysine (K)-specific demethylase 3A |
| chr4_-_59438633 | 1.21 |
ENSMUST00000040166.7
ENSMUST00000107544.1 |
Susd1
|
sushi domain containing 1 |
| chr10_-_130127055 | 1.16 |
ENSMUST00000074161.1
|
Olfr824
|
olfactory receptor 824 |
| chr2_-_7081207 | 1.16 |
ENSMUST00000114923.2
ENSMUST00000182706.1 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr11_-_75169519 | 1.12 |
ENSMUST00000055619.4
|
Hic1
|
hypermethylated in cancer 1 |
| chrX_+_10485121 | 1.04 |
ENSMUST00000076354.6
ENSMUST00000115526.1 |
Tspan7
|
tetraspanin 7 |
| chr10_+_116177217 | 0.93 |
ENSMUST00000148731.1
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr10_+_5593718 | 0.91 |
ENSMUST00000051809.8
|
Myct1
|
myc target 1 |
| chr3_-_146839365 | 0.90 |
ENSMUST00000084614.3
|
Gm10288
|
predicted gene 10288 |
| chr8_+_105636509 | 0.86 |
ENSMUST00000005841.9
|
Ctcf
|
CCCTC-binding factor |
| chr2_-_7081256 | 0.83 |
ENSMUST00000183209.1
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr10_-_13388830 | 0.81 |
ENSMUST00000079698.5
|
Phactr2
|
phosphatase and actin regulator 2 |
| chrX_-_95478107 | 0.78 |
ENSMUST00000033549.2
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr9_-_44713196 | 0.75 |
ENSMUST00000144251.1
ENSMUST00000156918.1 |
Phldb1
|
pleckstrin homology-like domain, family B, member 1 |
| chr2_+_32621750 | 0.72 |
ENSMUST00000113278.2
|
Ak1
|
adenylate kinase 1 |
| chr6_+_129397297 | 0.71 |
ENSMUST00000032262.7
|
Clec1b
|
C-type lectin domain family 1, member b |
| chr18_-_43477764 | 0.70 |
ENSMUST00000057110.9
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr7_+_45413657 | 0.60 |
ENSMUST00000058879.6
|
Ntf5
|
neurotrophin 5 |
| chr2_-_13011747 | 0.59 |
ENSMUST00000061545.5
|
C1ql3
|
C1q-like 3 |
| chr3_-_59262825 | 0.57 |
ENSMUST00000050360.7
|
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr7_-_44892358 | 0.52 |
ENSMUST00000003049.6
|
Med25
|
mediator of RNA polymerase II transcription, subunit 25 homolog (yeast) |
| chr2_-_26246707 | 0.52 |
ENSMUST00000166349.1
|
C030048H21Rik
|
RIKEN cDNA C030048H21 gene |
| chr16_-_29544852 | 0.45 |
ENSMUST00000039090.8
|
Atp13a4
|
ATPase type 13A4 |
| chr5_-_83355120 | 0.45 |
ENSMUST00000053543.4
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr8_+_74872692 | 0.42 |
ENSMUST00000034034.3
|
Isx
|
intestine specific homeobox |
| chr6_+_24597754 | 0.39 |
ENSMUST00000031694.6
|
Lmod2
|
leiomodin 2 (cardiac) |
| chr6_-_132957919 | 0.38 |
ENSMUST00000082085.4
|
Tas2r131
|
taste receptor, type 2, member 131 |
| chr8_-_4325096 | 0.35 |
ENSMUST00000098950.4
|
Elavl1
|
ELAV (embryonic lethal, abnormal vision)-like 1 (Hu antigen R) |
| chr15_-_77153772 | 0.33 |
ENSMUST00000166610.1
ENSMUST00000111581.2 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr3_+_90072641 | 0.32 |
ENSMUST00000121503.1
ENSMUST00000119570.1 ENSMUST00000062193.9 |
Tpm3
|
tropomyosin 3, gamma |
| chr2_-_60722636 | 0.32 |
ENSMUST00000028348.2
ENSMUST00000112517.1 |
Itgb6
|
integrin beta 6 |
| chrX_-_134111708 | 0.30 |
ENSMUST00000159259.1
ENSMUST00000113275.3 |
Nox1
|
NADPH oxidase 1 |
| chr4_-_14621805 | 0.29 |
ENSMUST00000042221.7
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr19_+_38264761 | 0.28 |
ENSMUST00000087252.5
|
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr11_-_61453992 | 0.27 |
ENSMUST00000060255.7
ENSMUST00000054927.7 ENSMUST00000102661.3 |
Rnf112
|
ring finger protein 112 |
| chr1_+_172341197 | 0.22 |
ENSMUST00000056136.3
|
Kcnj10
|
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr3_+_79884931 | 0.20 |
ENSMUST00000135021.1
|
Fam198b
|
family with sequence similarity 198, member B |
| chr12_+_95692212 | 0.19 |
ENSMUST00000057324.3
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr9_-_77251829 | 0.18 |
ENSMUST00000184322.1
ENSMUST00000184316.1 |
Mlip
|
muscular LMNA-interacting protein |
| chr5_+_64159429 | 0.15 |
ENSMUST00000043893.6
|
Tbc1d1
|
TBC1 domain family, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gata2_Gata1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.3 | 57.9 | GO:0002215 | defense response to nematode(GO:0002215) |
| 15.0 | 45.1 | GO:0030221 | basophil differentiation(GO:0030221) |
| 13.2 | 79.4 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 12.8 | 38.5 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 12.7 | 38.0 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 12.3 | 98.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 9.5 | 28.4 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 9.3 | 28.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 8.8 | 52.8 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 8.0 | 47.8 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 7.4 | 66.9 | GO:0002432 | granuloma formation(GO:0002432) |
| 7.4 | 51.7 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 5.6 | 44.4 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 4.5 | 44.9 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 3.9 | 38.9 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 3.8 | 19.1 | GO:2000471 | immunoglobulin biosynthetic process(GO:0002378) regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 3.8 | 30.4 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 3.3 | 19.8 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 2.9 | 37.1 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 2.8 | 33.7 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 2.5 | 12.7 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 2.5 | 32.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 2.4 | 19.0 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 2.3 | 16.2 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 2.2 | 31.3 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 2.2 | 26.8 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 2.2 | 22.2 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 2.2 | 38.9 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 2.2 | 21.5 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 2.1 | 49.0 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 2.1 | 10.5 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 1.9 | 9.3 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 1.6 | 1.6 | GO:0060529 | squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 1.6 | 61.1 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 1.6 | 32.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 1.6 | 23.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 1.5 | 13.9 | GO:0032264 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 1.5 | 37.3 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 1.3 | 7.7 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 1.2 | 14.9 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.9 | 2.8 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.9 | 11.1 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.9 | 23.0 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.9 | 3.4 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.8 | 12.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.8 | 37.8 | GO:0032094 | response to food(GO:0032094) |
| 0.8 | 1.6 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.7 | 44.7 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.6 | 5.7 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.6 | 7.2 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) |
| 0.6 | 7.7 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.5 | 2.9 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.5 | 2.8 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.5 | 1.4 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.4 | 10.4 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.4 | 1.6 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.4 | 2.9 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.4 | 3.2 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.3 | 3.7 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.3 | 1.3 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.3 | 22.2 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.3 | 2.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.3 | 5.5 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.3 | 18.5 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.3 | 1.6 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.3 | 4.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.3 | 17.6 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.3 | 2.3 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.2 | 18.0 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.2 | 9.0 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.2 | 0.9 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.2 | 5.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 1.6 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 13.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 2.7 | GO:0030220 | platelet formation(GO:0030220) |
| 0.2 | 2.8 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.2 | 6.6 | GO:0030168 | platelet activation(GO:0030168) |
| 0.2 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.2 | 9.9 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 1.8 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 261.7 | GO:0006508 | proteolysis(GO:0006508) |
| 0.1 | 0.4 | GO:1901738 | regulation of vitamin A metabolic process(GO:1901738) |
| 0.1 | 1.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 0.7 | GO:0046103 | ADP biosynthetic process(GO:0006172) inosine biosynthetic process(GO:0046103) |
| 0.1 | 11.5 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.1 | 0.7 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 4.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 3.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 18.9 | GO:0006887 | exocytosis(GO:0006887) |
| 0.1 | 0.2 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.1 | 2.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.4 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.3 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 3.3 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.5 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.4 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.3 | 98.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 6.6 | 72.9 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 3.7 | 37.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 3.1 | 9.3 | GO:0043512 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin A complex(GO:0043512) |
| 2.9 | 32.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 1.4 | 44.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.8 | 30.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.6 | 4.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.5 | 22.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.5 | 12.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.5 | 10.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.4 | 74.4 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.4 | 1.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.3 | 15.9 | GO:0002102 | podosome(GO:0002102) |
| 0.3 | 2.3 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.2 | 56.3 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.2 | 81.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 18.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 17.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 1.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.2 | 1.4 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.2 | 283.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.2 | 18.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 35.5 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 5.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 13.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 10.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 49.6 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.1 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 15.9 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 2.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.2 | GO:0071437 | invadopodium(GO:0071437) invadopodium membrane(GO:0071438) |
| 0.1 | 0.7 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 4.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 11.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.2 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 40.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 31.0 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 2.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 75.1 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 2.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.6 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 4.8 | GO:0000790 | nuclear chromatin(GO:0000790) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.3 | 98.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 11.3 | 79.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 7.4 | 22.2 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 6.3 | 38.0 | GO:0015265 | urea channel activity(GO:0015265) |
| 6.3 | 69.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 5.7 | 28.4 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 4.9 | 44.4 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 4.8 | 24.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 4.4 | 22.0 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 4.2 | 21.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 4.1 | 132.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 3.8 | 30.4 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 3.7 | 11.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 3.1 | 28.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 2.8 | 45.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 2.7 | 32.1 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 2.5 | 14.9 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 2.0 | 16.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 1.5 | 13.9 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.3 | 38.5 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 1.1 | 8.0 | GO:0016936 | galactoside binding(GO:0016936) |
| 1.1 | 3.3 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 1.0 | 47.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 1.0 | 9.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.8 | 12.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.8 | 23.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.8 | 8.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.8 | 167.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.7 | 30.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.7 | 13.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.6 | 43.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.6 | 3.7 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.6 | 6.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.6 | 19.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.5 | 38.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.5 | 68.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.5 | 7.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.5 | 26.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.5 | 1.4 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.4 | 1.8 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.4 | 45.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.4 | 3.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.4 | 13.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.3 | 66.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.3 | 2.8 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.3 | 7.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 1.6 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.3 | 12.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 26.6 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.2 | 13.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 16.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 2.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 37.1 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.2 | 1.6 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.2 | 9.0 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 43.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 14.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 10.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 1.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 2.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.6 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 3.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 4.9 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 28.4 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.1 | 0.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 5.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 2.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 1.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 2.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 57.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 1.2 | 40.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.9 | 22.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.9 | 69.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.9 | 13.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.8 | 66.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.5 | 19.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.4 | 28.0 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.3 | 15.4 | PID EPO PATHWAY | EPO signaling pathway |
| 0.2 | 23.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 16.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 20.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 54.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 3.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 7.2 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 2.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.4 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 3.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 6.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 28.6 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 1.5 | 44.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 1.4 | 28.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 1.3 | 41.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 1.2 | 10.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.9 | 13.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.9 | 21.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.9 | 41.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.9 | 19.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.8 | 10.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.8 | 39.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.7 | 13.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.7 | 29.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.7 | 13.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.5 | 5.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.4 | 5.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.4 | 60.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.3 | 6.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.3 | 6.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 3.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 7.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 18.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 4.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 5.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |