Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Gata4
Z-value: 2.44
Transcription factors associated with Gata4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gata4
|
ENSMUSG00000021944.9 | GATA binding protein 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gata4 | mm10_v2_chr14_-_63271682_63271702 | 0.32 | 5.9e-02 | Click! |
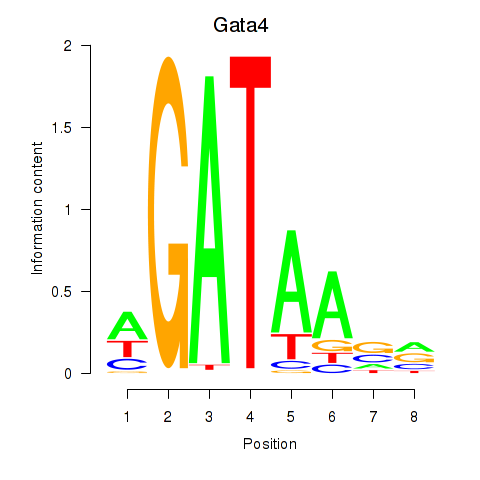
Activity profile of Gata4 motif
Sorted Z-values of Gata4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_26199008 | 46.61 |
ENSMUST00000142410.1
ENSMUST00000120333.1 ENSMUST00000039113.7 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr6_-_41314700 | 28.39 |
ENSMUST00000064324.5
|
Try5
|
trypsin 5 |
| chr6_+_30639218 | 26.12 |
ENSMUST00000031806.9
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr2_+_84980458 | 23.24 |
ENSMUST00000028467.5
|
Prg2
|
proteoglycan 2, bone marrow |
| chr8_-_85380964 | 23.11 |
ENSMUST00000122452.1
|
Mylk3
|
myosin light chain kinase 3 |
| chr19_+_58759700 | 21.97 |
ENSMUST00000026081.3
|
Pnliprp2
|
pancreatic lipase-related protein 2 |
| chr6_+_41354105 | 21.40 |
ENSMUST00000072103.5
|
Try10
|
trypsin 10 |
| chr4_-_137430517 | 21.09 |
ENSMUST00000102522.4
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr11_+_32276400 | 20.62 |
ENSMUST00000020531.2
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr4_-_46404224 | 20.43 |
ENSMUST00000107764.2
|
Hemgn
|
hemogen |
| chr6_+_41302265 | 19.39 |
ENSMUST00000031913.4
|
Try4
|
trypsin 4 |
| chr5_-_73191848 | 19.32 |
ENSMUST00000176910.1
|
Fryl
|
furry homolog-like (Drosophila) |
| chr4_+_34893772 | 18.22 |
ENSMUST00000029975.3
ENSMUST00000135871.1 ENSMUST00000108130.1 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr17_-_35085609 | 18.13 |
ENSMUST00000038507.6
|
Ly6g6f
|
lymphocyte antigen 6 complex, locus G6F |
| chr11_+_32276893 | 17.98 |
ENSMUST00000145569.1
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr2_-_121036750 | 17.12 |
ENSMUST00000023987.5
|
Epb4.2
|
erythrocyte protein band 4.2 |
| chr11_-_83286722 | 17.09 |
ENSMUST00000163961.2
|
Slfn14
|
schlafen family member 14 |
| chr17_-_35066170 | 16.83 |
ENSMUST00000174190.1
ENSMUST00000097337.1 |
AU023871
|
expressed sequence AU023871 |
| chr17_+_36869567 | 16.65 |
ENSMUST00000060524.9
|
Trim10
|
tripartite motif-containing 10 |
| chr17_-_28560704 | 16.63 |
ENSMUST00000114785.1
ENSMUST00000025062.3 |
Clps
|
colipase, pancreatic |
| chr6_+_30541582 | 16.57 |
ENSMUST00000096066.4
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr7_+_110773658 | 16.11 |
ENSMUST00000143786.1
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr3_-_20242173 | 15.28 |
ENSMUST00000001921.1
|
Cpa3
|
carboxypeptidase A3, mast cell |
| chr2_-_121037048 | 15.03 |
ENSMUST00000102490.3
|
Epb4.2
|
erythrocyte protein band 4.2 |
| chr1_-_132367879 | 15.02 |
ENSMUST00000142609.1
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr5_+_115466234 | 14.47 |
ENSMUST00000145785.1
ENSMUST00000031495.4 ENSMUST00000112071.1 ENSMUST00000125568.1 |
Pla2g1b
|
phospholipase A2, group IB, pancreas |
| chr9_+_7558429 | 14.39 |
ENSMUST00000018765.2
|
Mmp8
|
matrix metallopeptidase 8 |
| chr4_-_137409777 | 14.23 |
ENSMUST00000024200.6
|
Gm13011
|
predicted gene 13011 |
| chr7_+_28540863 | 14.04 |
ENSMUST00000119180.2
|
Sycn
|
syncollin |
| chr3_-_84479418 | 13.09 |
ENSMUST00000091002.1
|
Fhdc1
|
FH2 domain containing 1 |
| chr17_+_31208049 | 12.95 |
ENSMUST00000173776.1
|
Ubash3a
|
ubiquitin associated and SH3 domain containing, A |
| chrX_+_93654863 | 12.58 |
ENSMUST00000113933.2
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr3_+_108364882 | 12.58 |
ENSMUST00000090563.5
|
Mybphl
|
myosin binding protein H-like |
| chr11_-_116076986 | 12.39 |
ENSMUST00000153408.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chrX_+_93675088 | 12.37 |
ENSMUST00000045898.3
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr1_+_174172738 | 11.61 |
ENSMUST00000027817.7
|
Spta1
|
spectrin alpha, erythrocytic 1 |
| chr10_-_62342674 | 11.48 |
ENSMUST00000143179.1
ENSMUST00000130422.1 |
Hk1
|
hexokinase 1 |
| chr8_+_84701430 | 11.19 |
ENSMUST00000037165.4
|
Lyl1
|
lymphoblastomic leukemia 1 |
| chr1_-_171281181 | 11.05 |
ENSMUST00000073120.4
|
Ppox
|
protoporphyrinogen oxidase |
| chr9_+_21029373 | 10.99 |
ENSMUST00000001040.5
|
Icam4
|
intercellular adhesion molecule 4, Landsteiner-Wiener blood group |
| chr4_-_87806296 | 10.59 |
ENSMUST00000126353.1
ENSMUST00000149357.1 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr15_-_103251465 | 10.33 |
ENSMUST00000133600.1
ENSMUST00000134554.1 ENSMUST00000156927.1 ENSMUST00000149111.1 ENSMUST00000132836.1 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr6_-_129507107 | 10.28 |
ENSMUST00000183258.1
ENSMUST00000182784.1 ENSMUST00000032265.6 ENSMUST00000162815.1 |
Olr1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr16_+_38362205 | 10.03 |
ENSMUST00000023494.6
|
Popdc2
|
popeye domain containing 2 |
| chr4_-_87806276 | 9.68 |
ENSMUST00000148059.1
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr4_+_119637704 | 9.44 |
ENSMUST00000024015.2
|
Guca2a
|
guanylate cyclase activator 2a (guanylin) |
| chr7_-_99238564 | 8.84 |
ENSMUST00000064231.7
|
Mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr16_+_38362234 | 8.72 |
ENSMUST00000114739.1
|
Popdc2
|
popeye domain containing 2 |
| chr6_-_122609964 | 8.29 |
ENSMUST00000032211.4
|
Gdf3
|
growth differentiation factor 3 |
| chr2_+_103957976 | 8.22 |
ENSMUST00000156813.1
ENSMUST00000170926.1 |
Lmo2
|
LIM domain only 2 |
| chr9_-_21963568 | 7.94 |
ENSMUST00000006397.5
|
Epor
|
erythropoietin receptor |
| chr19_-_10678001 | 7.84 |
ENSMUST00000025647.5
|
Pga5
|
pepsinogen 5, group I |
| chrX_-_8145713 | 7.69 |
ENSMUST00000115615.2
ENSMUST00000115616.1 ENSMUST00000115621.2 |
Rbm3
|
RNA binding motif protein 3 |
| chr10_-_80421847 | 7.22 |
ENSMUST00000156244.1
|
Tcf3
|
transcription factor 3 |
| chr3_+_103832562 | 7.05 |
ENSMUST00000062945.5
|
Bcl2l15
|
BCLl2-like 15 |
| chr11_-_102469839 | 6.91 |
ENSMUST00000103086.3
|
Itga2b
|
integrin alpha 2b |
| chr13_-_73700721 | 6.40 |
ENSMUST00000022048.5
|
Slc6a19
|
solute carrier family 6 (neurotransmitter transporter), member 19 |
| chr11_-_116077954 | 6.39 |
ENSMUST00000106451.1
ENSMUST00000075036.2 |
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr11_-_116077606 | 6.35 |
ENSMUST00000106450.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr13_-_23465872 | 6.28 |
ENSMUST00000041674.7
|
Btn1a1
|
butyrophilin, subfamily 1, member A1 |
| chr4_+_13751297 | 6.21 |
ENSMUST00000105566.2
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr4_+_40920047 | 6.21 |
ENSMUST00000030122.4
|
Spink4
|
serine peptidase inhibitor, Kazal type 4 |
| chr11_-_116077562 | 6.20 |
ENSMUST00000174822.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr11_-_116077927 | 6.09 |
ENSMUST00000156545.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr2_+_14873656 | 5.93 |
ENSMUST00000114718.1
ENSMUST00000114719.1 |
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chrX_-_107403295 | 5.84 |
ENSMUST00000033591.5
|
Itm2a
|
integral membrane protein 2A |
| chr7_+_99535439 | 5.78 |
ENSMUST00000098266.2
ENSMUST00000179755.1 |
Arrb1
|
arrestin, beta 1 |
| chr8_-_46211284 | 5.54 |
ENSMUST00000034049.4
|
Slc25a4
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 |
| chr4_-_43558386 | 5.48 |
ENSMUST00000130353.1
|
Tln1
|
talin 1 |
| chr3_-_107221722 | 5.44 |
ENSMUST00000029504.8
|
Cym
|
chymosin |
| chr16_-_92826004 | 5.14 |
ENSMUST00000023673.7
|
Runx1
|
runt related transcription factor 1 |
| chr5_-_24329556 | 5.12 |
ENSMUST00000115098.2
|
Kcnh2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr11_-_99322943 | 5.07 |
ENSMUST00000038004.2
|
Krt25
|
keratin 25 |
| chr2_+_4559742 | 5.00 |
ENSMUST00000176828.1
|
Frmd4a
|
FERM domain containing 4A |
| chr11_+_3330781 | 4.94 |
ENSMUST00000136536.1
ENSMUST00000093399.4 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr2_-_153241402 | 4.93 |
ENSMUST00000056924.7
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr15_+_57985873 | 4.76 |
ENSMUST00000050374.2
|
Fam83a
|
family with sequence similarity 83, member A |
| chr2_+_72476225 | 4.61 |
ENSMUST00000157019.1
|
Cdca7
|
cell division cycle associated 7 |
| chr11_+_101260569 | 4.53 |
ENSMUST00000103108.1
|
Wnk4
|
WNK lysine deficient protein kinase 4 |
| chr13_-_62858364 | 4.46 |
ENSMUST00000021907.7
|
Fbp2
|
fructose bisphosphatase 2 |
| chr2_+_72476159 | 4.39 |
ENSMUST00000102691.4
|
Cdca7
|
cell division cycle associated 7 |
| chr13_-_23465895 | 4.35 |
ENSMUST00000110434.1
|
Btn1a1
|
butyrophilin, subfamily 1, member A1 |
| chr1_+_12718496 | 4.16 |
ENSMUST00000088585.3
|
Sulf1
|
sulfatase 1 |
| chr11_+_3330401 | 3.86 |
ENSMUST00000045153.4
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr1_-_173333503 | 3.70 |
ENSMUST00000038227.4
|
Darc
|
Duffy blood group, chemokine receptor |
| chr9_-_44342332 | 3.68 |
ENSMUST00000097558.3
|
Hmbs
|
hydroxymethylbilane synthase |
| chr10_-_80433615 | 3.67 |
ENSMUST00000105346.3
ENSMUST00000020377.6 ENSMUST00000105340.1 ENSMUST00000020379.6 ENSMUST00000105344.1 ENSMUST00000105342.1 ENSMUST00000105345.3 ENSMUST00000105343.1 |
Tcf3
|
transcription factor 3 |
| chr14_-_54966570 | 3.60 |
ENSMUST00000124930.1
ENSMUST00000134256.1 ENSMUST00000081857.6 ENSMUST00000145322.1 |
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr6_-_136922169 | 3.58 |
ENSMUST00000032343.6
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr7_+_28833975 | 3.53 |
ENSMUST00000066723.8
|
Lgals4
|
lectin, galactose binding, soluble 4 |
| chr11_+_70505244 | 3.32 |
ENSMUST00000019063.2
|
Tm4sf5
|
transmembrane 4 superfamily member 5 |
| chr3_-_102964124 | 3.29 |
ENSMUST00000058899.8
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr2_+_74704615 | 3.26 |
ENSMUST00000151380.1
|
Hoxd8
|
homeobox D8 |
| chr11_-_6230127 | 3.17 |
ENSMUST00000004505.2
|
Npc1l1
|
NPC1-like 1 |
| chr6_+_87428986 | 3.16 |
ENSMUST00000032125.5
|
Bmp10
|
bone morphogenetic protein 10 |
| chrX_+_56779699 | 3.13 |
ENSMUST00000114772.2
ENSMUST00000114768.3 ENSMUST00000155882.1 |
Fhl1
|
four and a half LIM domains 1 |
| chr2_-_59948155 | 2.94 |
ENSMUST00000153136.1
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr17_-_31144271 | 2.91 |
ENSMUST00000024826.7
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr15_-_102231920 | 2.90 |
ENSMUST00000001327.3
ENSMUST00000127014.1 |
Itgb7
|
integrin beta 7 |
| chrX_+_159840463 | 2.89 |
ENSMUST00000112451.1
ENSMUST00000112453.2 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chrX_+_150547375 | 2.89 |
ENSMUST00000066337.6
ENSMUST00000112715.1 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr2_-_33087169 | 2.88 |
ENSMUST00000102810.3
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr3_-_14808358 | 2.86 |
ENSMUST00000181860.1
ENSMUST00000144327.2 |
Car1
|
carbonic anhydrase 1 |
| chr6_+_125552948 | 2.83 |
ENSMUST00000112254.1
ENSMUST00000112253.1 ENSMUST00000001995.7 |
Vwf
|
Von Willebrand factor homolog |
| chr2_+_164403194 | 2.81 |
ENSMUST00000017151.1
|
Rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr17_-_33713372 | 2.65 |
ENSMUST00000173392.1
|
March2
|
membrane-associated ring finger (C3HC4) 2 |
| chr14_+_75455957 | 2.64 |
ENSMUST00000164848.1
|
Siah3
|
seven in absentia homolog 3 (Drosophila) |
| chr11_+_98386450 | 2.58 |
ENSMUST00000041301.7
|
Pnmt
|
phenylethanolamine-N-methyltransferase |
| chr2_+_74704861 | 2.57 |
ENSMUST00000019749.3
|
Hoxd8
|
homeobox D8 |
| chr3_+_98222148 | 2.51 |
ENSMUST00000029469.4
|
Reg4
|
regenerating islet-derived family, member 4 |
| chr11_-_57832679 | 2.48 |
ENSMUST00000160392.2
ENSMUST00000108845.2 |
Hand1
|
heart and neural crest derivatives expressed transcript 1 |
| chrX_+_56779437 | 2.43 |
ENSMUST00000114773.3
|
Fhl1
|
four and a half LIM domains 1 |
| chr10_+_80019621 | 2.36 |
ENSMUST00000043311.6
|
Hmha1
|
histocompatibility (minor) HA-1 |
| chr3_-_144819494 | 2.28 |
ENSMUST00000029929.7
|
Clca2
|
chloride channel calcium activated 2 |
| chr3_-_129755305 | 2.24 |
ENSMUST00000029653.2
|
Egf
|
epidermal growth factor |
| chr18_-_61707583 | 2.24 |
ENSMUST00000025472.1
|
Pcyox1l
|
prenylcysteine oxidase 1 like |
| chr3_-_63851251 | 2.19 |
ENSMUST00000162269.2
ENSMUST00000159676.2 ENSMUST00000175947.1 |
Plch1
|
phospholipase C, eta 1 |
| chr1_-_160153571 | 2.16 |
ENSMUST00000039178.5
|
Tnn
|
tenascin N |
| chr7_-_98112618 | 2.15 |
ENSMUST00000153657.1
|
Myo7a
|
myosin VIIA |
| chr1_+_179803376 | 2.14 |
ENSMUST00000097454.2
|
Gm10518
|
predicted gene 10518 |
| chr6_+_129397478 | 2.13 |
ENSMUST00000112081.2
ENSMUST00000112079.2 |
Clec1b
|
C-type lectin domain family 1, member b |
| chr11_+_4986824 | 2.09 |
ENSMUST00000009234.9
ENSMUST00000109897.1 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr3_+_79884576 | 1.92 |
ENSMUST00000145992.1
|
Fam198b
|
family with sequence similarity 198, member B |
| chr4_+_150148905 | 1.86 |
ENSMUST00000059893.7
|
Slc2a7
|
solute carrier family 2 (facilitated glucose transporter), member 7 |
| chr16_+_57353093 | 1.85 |
ENSMUST00000159816.1
|
Filip1l
|
filamin A interacting protein 1-like |
| chr16_-_63864114 | 1.84 |
ENSMUST00000064405.6
|
Epha3
|
Eph receptor A3 |
| chr12_-_91746020 | 1.64 |
ENSMUST00000166967.1
|
Ston2
|
stonin 2 |
| chr4_-_32923455 | 1.55 |
ENSMUST00000035719.4
ENSMUST00000084749.1 |
Ankrd6
|
ankyrin repeat domain 6 |
| chr15_+_76904070 | 1.45 |
ENSMUST00000004072.8
|
Rpl8
|
ribosomal protein L8 |
| chr7_-_135528645 | 1.44 |
ENSMUST00000053716.7
|
Clrn3
|
clarin 3 |
| chr3_+_96596628 | 1.39 |
ENSMUST00000058943.7
|
Ankrd34a
|
ankyrin repeat domain 34A |
| chr8_-_122699066 | 1.36 |
ENSMUST00000127984.1
|
Cbfa2t3
|
core-binding factor, runt domain, alpha subunit 2, translocated to, 3 (human) |
| chr2_-_33086366 | 1.33 |
ENSMUST00000049618.2
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr10_-_19011948 | 1.32 |
ENSMUST00000105527.1
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chrX_-_7572843 | 1.28 |
ENSMUST00000132788.1
|
Ppp1r3f
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F |
| chr6_-_124768330 | 1.23 |
ENSMUST00000135626.1
|
Eno2
|
enolase 2, gamma neuronal |
| chr9_+_66350465 | 1.17 |
ENSMUST00000042824.6
|
Herc1
|
hect (homologous to the E6-AP (UBE3A) carboxyl terminus) domain and RCC1 (CHC1)-like domain (RLD) 1 |
| chr10_+_116177351 | 1.09 |
ENSMUST00000155606.1
ENSMUST00000128399.1 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr1_-_4496363 | 1.05 |
ENSMUST00000116652.2
|
Sox17
|
SRY-box containing gene 17 |
| chrX_+_10485121 | 1.01 |
ENSMUST00000076354.6
ENSMUST00000115526.1 |
Tspan7
|
tetraspanin 7 |
| chr3_-_59262825 | 1.00 |
ENSMUST00000050360.7
|
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr9_-_106887000 | 0.89 |
ENSMUST00000055843.7
|
Rbm15b
|
RNA binding motif protein 15B |
| chr3_-_144849301 | 0.88 |
ENSMUST00000159989.1
|
Clca4
|
chloride channel calcium activated 4 |
| chr2_+_71786923 | 0.87 |
ENSMUST00000112101.1
ENSMUST00000028522.3 |
Itga6
|
integrin alpha 6 |
| chr18_-_24020307 | 0.70 |
ENSMUST00000153337.1
ENSMUST00000148525.1 |
Zfp191
|
zinc finger protein 191 |
| chr17_+_57249450 | 0.67 |
ENSMUST00000019631.9
|
Trip10
|
thyroid hormone receptor interactor 10 |
| chr12_-_69893162 | 0.66 |
ENSMUST00000049239.7
ENSMUST00000110570.1 |
Map4k5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr6_+_129397297 | 0.65 |
ENSMUST00000032262.7
|
Clec1b
|
C-type lectin domain family 1, member b |
| chr12_+_95692212 | 0.63 |
ENSMUST00000057324.3
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr10_+_69219357 | 0.60 |
ENSMUST00000172261.1
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr2_-_26246707 | 0.53 |
ENSMUST00000166349.1
|
C030048H21Rik
|
RIKEN cDNA C030048H21 gene |
| chr3_+_79884496 | 0.52 |
ENSMUST00000118853.1
|
Fam198b
|
family with sequence similarity 198, member B |
| chr8_+_74872692 | 0.51 |
ENSMUST00000034034.3
|
Isx
|
intestine specific homeobox |
| chr13_-_22219820 | 0.49 |
ENSMUST00000057516.1
|
Vmn1r193
|
vomeronasal 1 receptor 193 |
| chr10_+_76147451 | 0.48 |
ENSMUST00000020450.3
|
Slc5a4a
|
solute carrier family 5, member 4a |
| chr1_-_4496400 | 0.45 |
ENSMUST00000027035.3
|
Sox17
|
SRY-box containing gene 17 |
| chr3_+_59952185 | 0.43 |
ENSMUST00000094227.3
|
Gm9696
|
predicted gene 9696 |
| chrX_-_95478107 | 0.42 |
ENSMUST00000033549.2
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr7_-_44892358 | 0.40 |
ENSMUST00000003049.6
|
Med25
|
mediator of RNA polymerase II transcription, subunit 25 homolog (yeast) |
| chr7_+_45413657 | 0.37 |
ENSMUST00000058879.6
|
Ntf5
|
neurotrophin 5 |
| chr15_-_77153772 | 0.36 |
ENSMUST00000166610.1
ENSMUST00000111581.2 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr11_+_88047693 | 0.35 |
ENSMUST00000079866.4
|
Srsf1
|
serine/arginine-rich splicing factor 1 |
| chr10_+_39369750 | 0.34 |
ENSMUST00000063091.6
ENSMUST00000099967.3 ENSMUST00000126486.1 |
Fyn
|
Fyn proto-oncogene |
| chr6_+_17306415 | 0.32 |
ENSMUST00000150901.1
|
Cav1
|
caveolin 1, caveolae protein |
| chr2_+_131133497 | 0.31 |
ENSMUST00000110225.1
|
Gm11037
|
predicted gene 11037 |
| chr15_+_60822947 | 0.31 |
ENSMUST00000180730.1
|
9930014A18Rik
|
RIKEN cDNA 9930014A18 gene |
| chr10_+_39133981 | 0.29 |
ENSMUST00000019991.7
|
Tube1
|
epsilon-tubulin 1 |
| chr18_-_43477764 | 0.27 |
ENSMUST00000057110.9
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chrX_+_73757069 | 0.23 |
ENSMUST00000002079.6
|
Plxnb3
|
plexin B3 |
| chrX_+_112311334 | 0.20 |
ENSMUST00000026599.3
ENSMUST00000113415.1 |
Apool
|
apolipoprotein O-like |
| chr11_-_99351086 | 0.19 |
ENSMUST00000017732.2
|
Krt27
|
keratin 27 |
| chr8_-_4325096 | 0.16 |
ENSMUST00000098950.4
|
Elavl1
|
ELAV (embryonic lethal, abnormal vision)-like 1 (Hu antigen R) |
| chr1_-_14310198 | 0.14 |
ENSMUST00000168081.2
ENSMUST00000027066.6 |
Eya1
|
eyes absent 1 homolog (Drosophila) |
| chr2_-_7081207 | 0.12 |
ENSMUST00000114923.2
ENSMUST00000182706.1 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr2_+_65845767 | 0.11 |
ENSMUST00000122912.1
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr10_-_83648631 | 0.10 |
ENSMUST00000146876.2
ENSMUST00000176294.1 |
Appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr1_-_107052331 | 0.09 |
ENSMUST00000112717.1
|
Serpinb3a
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3A |
| chr11_+_88047788 | 0.09 |
ENSMUST00000107920.3
|
Srsf1
|
serine/arginine-rich splicing factor 1 |
| chrX_+_169685191 | 0.08 |
ENSMUST00000112104.1
ENSMUST00000112107.1 |
Mid1
|
midline 1 |
| chr4_-_14621805 | 0.03 |
ENSMUST00000042221.7
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr5_-_123666682 | 0.02 |
ENSMUST00000149410.1
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gata4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.7 | 23.2 | GO:0002215 | defense response to nematode(GO:0002215) |
| 4.8 | 38.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 4.8 | 14.5 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 4.3 | 38.7 | GO:0002432 | granuloma formation(GO:0002432) |
| 3.9 | 23.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 3.1 | 18.8 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 2.9 | 8.8 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 2.0 | 5.9 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 1.9 | 19.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 1.8 | 5.5 | GO:0015866 | ADP transport(GO:0015866) |
| 1.8 | 16.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 1.7 | 5.1 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 1.7 | 18.2 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 1.6 | 11.2 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 1.4 | 11.5 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 1.4 | 6.9 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 1.4 | 21.8 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 1.4 | 10.9 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 1.3 | 22.0 | GO:0019374 | galactolipid metabolic process(GO:0019374) glycolipid catabolic process(GO:0019377) |
| 1.3 | 5.1 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 1.3 | 8.8 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 1.2 | 15.3 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 1.1 | 4.5 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 1.1 | 42.3 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 1.0 | 4.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 1.0 | 5.8 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 1.0 | 5.8 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 1.0 | 8.7 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.9 | 17.1 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.9 | 10.3 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.8 | 2.5 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.8 | 10.6 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.7 | 2.2 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.7 | 11.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.7 | 16.6 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.7 | 5.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.7 | 2.6 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.6 | 13.0 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.6 | 1.8 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.6 | 14.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.5 | 8.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.5 | 2.2 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.5 | 7.9 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.5 | 2.6 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.5 | 4.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.5 | 1.5 | GO:0060796 | stem cell fate specification(GO:0048866) regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) regulation of cardiac cell fate specification(GO:2000043) |
| 0.5 | 18.2 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.5 | 2.7 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.4 | 2.9 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.4 | 16.6 | GO:0032094 | response to food(GO:0032094) |
| 0.4 | 2.2 | GO:0007262 | STAT protein import into nucleus(GO:0007262) negative regulation of cholesterol efflux(GO:0090370) |
| 0.4 | 24.9 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.4 | 4.9 | GO:0034377 | plasma lipoprotein particle assembly(GO:0034377) |
| 0.3 | 15.0 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.3 | 1.0 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 | 2.2 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.2 | 1.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 1.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 5.1 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.2 | 0.5 | GO:1901738 | regulation of vitamin A metabolic process(GO:1901738) |
| 0.2 | 7.7 | GO:0009409 | response to cold(GO:0009409) production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.2 | 6.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.2 | 1.4 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 5.6 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 0.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 2.8 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 5.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 6.2 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 1.3 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 6.9 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 10.5 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 0.9 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 2.8 | GO:0030168 | platelet activation(GO:0030168) |
| 0.1 | 2.9 | GO:0072678 | T cell migration(GO:0072678) |
| 0.1 | 0.6 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.3 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 0.7 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.1 | 2.9 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 8.4 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.1 | 0.2 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.1 | 1.2 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.2 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 6.5 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 1.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 4.0 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 1.6 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 14.0 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 61.8 | GO:0006508 | proteolysis(GO:0006508) |
| 0.0 | 0.5 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 5.8 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 2.2 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 1.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 2.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.2 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 38.6 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 3.6 | 39.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 1.9 | 11.6 | GO:0032437 | cuticular plate(GO:0032437) |
| 1.5 | 22.0 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 1.3 | 5.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.7 | 5.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.5 | 13.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.4 | 20.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.4 | 11.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.3 | 10.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 2.2 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.2 | 10.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.2 | 7.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.2 | 3.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 14.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 1.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 2.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 16.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 18.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 156.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 9.1 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.1 | 9.5 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 9.6 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 12.6 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 5.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.6 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 1.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 3.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 5.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 5.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 3.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 6.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.3 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 3.7 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 6.3 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 2.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 24.5 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 45.3 | GO:0005576 | extracellular region(GO:0005576) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 24.9 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 4.8 | 38.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 4.2 | 46.6 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 3.3 | 23.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 2.2 | 8.8 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 1.9 | 5.8 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 1.9 | 9.4 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 1.8 | 58.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 1.8 | 16.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.8 | 8.8 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 1.6 | 14.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 1.5 | 22.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 1.4 | 6.9 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 1.4 | 10.9 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 1.3 | 11.5 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 1.2 | 5.9 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 1.0 | 2.9 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.9 | 7.7 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.9 | 5.1 | GO:1902282 | phosphorelay sensor kinase activity(GO:0000155) voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.8 | 4.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.7 | 2.2 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.7 | 10.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.6 | 3.7 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.6 | 11.6 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.5 | 3.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.5 | 3.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.5 | 6.9 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.5 | 13.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.5 | 5.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.5 | 94.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.5 | 5.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.4 | 4.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.4 | 2.9 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 1.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.3 | 4.5 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.3 | 1.8 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.3 | 17.1 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.3 | 2.2 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.3 | 11.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.3 | 4.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 10.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 2.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 1.8 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 40.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.2 | 10.7 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 2.8 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.2 | 1.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.9 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 17.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 6.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 2.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 2.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 5.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 2.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 11.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.0 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 3.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 17.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 0.4 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.2 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 6.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 1.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 5.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 5.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 2.6 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 1.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 11.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 9.2 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 0.5 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 1.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.4 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 22.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.5 | 47.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.4 | 10.7 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.3 | 11.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 8.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 8.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 5.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 16.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 33.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 4.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 9.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.7 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 2.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 1.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 2.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 2.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 18.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.8 | 16.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.8 | 15.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.8 | 23.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.7 | 14.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.6 | 13.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.5 | 5.5 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.3 | 1.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.3 | 11.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 10.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 7.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 9.8 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.2 | 1.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 6.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 8.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 5.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 2.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 5.9 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 4.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 6.7 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 1.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 2.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |