Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
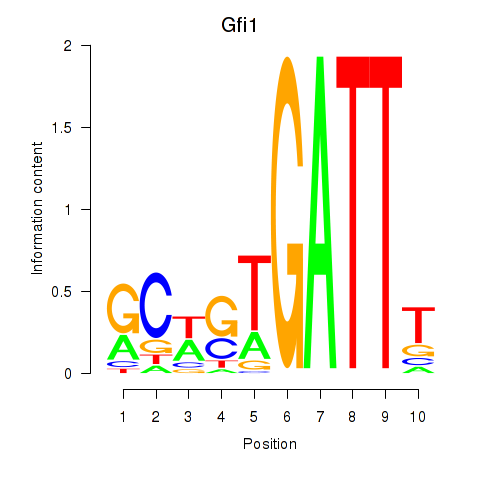
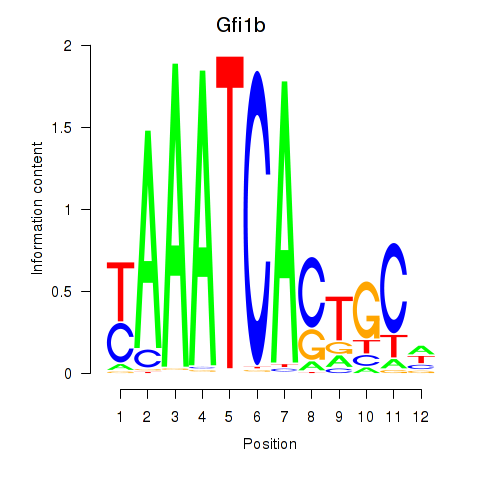
Results for Gfi1_Gfi1b
Z-value: 1.27
Transcription factors associated with Gfi1_Gfi1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gfi1
|
ENSMUSG00000029275.11 | growth factor independent 1 transcription repressor |
|
Gfi1b
|
ENSMUSG00000026815.8 | growth factor independent 1B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gfi1 | mm10_v2_chr5_-_107723954_107723974 | -0.81 | 1.5e-09 | Click! |
| Gfi1b | mm10_v2_chr2_-_28621932_28621982 | -0.81 | 2.1e-09 | Click! |
Activity profile of Gfi1_Gfi1b motif
Sorted Z-values of Gfi1_Gfi1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_127759780 | 10.76 |
ENSMUST00000128247.1
|
RP23-386P10.11
|
Protein Rdh9 |
| chr5_-_87091150 | 7.79 |
ENSMUST00000154455.1
|
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr6_+_121838514 | 7.34 |
ENSMUST00000032228.8
|
Mug1
|
murinoglobulin 1 |
| chr6_+_122006798 | 6.69 |
ENSMUST00000081777.6
|
Mug2
|
murinoglobulin 2 |
| chr8_+_46010596 | 6.01 |
ENSMUST00000110381.2
|
Lrp2bp
|
Lrp2 binding protein |
| chr15_+_6445320 | 5.45 |
ENSMUST00000022749.9
|
C9
|
complement component 9 |
| chr6_-_127109517 | 4.95 |
ENSMUST00000039913.8
|
9630033F20Rik
|
RIKEN cDNA 9630033F20 gene |
| chr10_-_127370535 | 4.75 |
ENSMUST00000026472.8
|
Inhbc
|
inhibin beta-C |
| chr5_+_90891234 | 4.61 |
ENSMUST00000031327.8
|
Cxcl1
|
chemokine (C-X-C motif) ligand 1 |
| chr1_+_167618246 | 4.49 |
ENSMUST00000111380.1
|
Rxrg
|
retinoid X receptor gamma |
| chr16_-_46010212 | 4.46 |
ENSMUST00000130481.1
|
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr10_-_128960965 | 4.37 |
ENSMUST00000026398.3
|
Mettl7b
|
methyltransferase like 7B |
| chr3_+_138415484 | 3.96 |
ENSMUST00000161312.1
ENSMUST00000013458.8 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr4_-_49408042 | 3.93 |
ENSMUST00000081541.2
|
Acnat2
|
acyl-coenzyme A amino acid N-acyltransferase 2 |
| chr14_+_55560904 | 3.91 |
ENSMUST00000072530.4
ENSMUST00000128490.1 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr3_-_98630309 | 3.86 |
ENSMUST00000044094.4
|
Hsd3b5
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 5 |
| chr11_+_101367542 | 3.81 |
ENSMUST00000019469.2
|
G6pc
|
glucose-6-phosphatase, catalytic |
| chr4_-_108118504 | 3.64 |
ENSMUST00000149106.1
|
Scp2
|
sterol carrier protein 2, liver |
| chr8_-_117673682 | 3.60 |
ENSMUST00000173522.1
ENSMUST00000174450.1 |
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr7_+_51878967 | 3.47 |
ENSMUST00000051912.6
|
Gas2
|
growth arrest specific 2 |
| chr1_+_60908993 | 3.37 |
ENSMUST00000027164.2
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr19_-_20727533 | 3.37 |
ENSMUST00000025656.3
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr1_+_93512079 | 3.36 |
ENSMUST00000120301.1
ENSMUST00000041983.4 ENSMUST00000122402.1 |
Farp2
|
FERM, RhoGEF and pleckstrin domain protein 2 |
| chr19_-_34877880 | 3.34 |
ENSMUST00000112460.1
|
Pank1
|
pantothenate kinase 1 |
| chr2_-_160872552 | 3.22 |
ENSMUST00000103111.2
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr7_+_51879041 | 3.21 |
ENSMUST00000107591.2
|
Gas2
|
growth arrest specific 2 |
| chr18_-_74961252 | 3.20 |
ENSMUST00000066532.4
|
Lipg
|
lipase, endothelial |
| chr10_-_109010955 | 3.16 |
ENSMUST00000105276.1
ENSMUST00000064054.7 |
Syt1
|
synaptotagmin I |
| chr4_-_108118528 | 3.01 |
ENSMUST00000030340.8
|
Scp2
|
sterol carrier protein 2, liver |
| chr16_+_38089001 | 2.97 |
ENSMUST00000023507.6
|
Gsk3b
|
glycogen synthase kinase 3 beta |
| chr8_-_61591130 | 2.97 |
ENSMUST00000135439.1
ENSMUST00000121200.1 |
Palld
|
palladin, cytoskeletal associated protein |
| chr1_-_169747634 | 2.94 |
ENSMUST00000027991.5
ENSMUST00000111357.1 |
Rgs4
|
regulator of G-protein signaling 4 |
| chr2_-_110305730 | 2.92 |
ENSMUST00000046233.2
|
Bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase) |
| chr12_-_98577940 | 2.87 |
ENSMUST00000110113.1
|
Kcnk10
|
potassium channel, subfamily K, member 10 |
| chr14_+_55560480 | 2.86 |
ENSMUST00000121622.1
ENSMUST00000143431.1 ENSMUST00000150481.1 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr8_-_84893887 | 2.78 |
ENSMUST00000003907.7
ENSMUST00000182458.1 ENSMUST00000109745.1 ENSMUST00000142748.1 |
Gcdh
|
glutaryl-Coenzyme A dehydrogenase |
| chr1_+_167598450 | 2.77 |
ENSMUST00000111386.1
ENSMUST00000111384.1 |
Rxrg
|
retinoid X receptor gamma |
| chr1_-_9962809 | 2.74 |
ENSMUST00000097824.2
|
Gm10567
|
predicted gene 10567 |
| chr11_+_112782182 | 2.71 |
ENSMUST00000000579.2
|
Sox9
|
SRY-box containing gene 9 |
| chr2_+_58754910 | 2.62 |
ENSMUST00000059102.6
|
Upp2
|
uridine phosphorylase 2 |
| chr19_+_12633507 | 2.61 |
ENSMUST00000119960.1
|
Glyat
|
glycine-N-acyltransferase |
| chr7_-_99626936 | 2.59 |
ENSMUST00000178124.1
|
Gm4980
|
predicted gene 4980 |
| chr13_-_71963713 | 2.59 |
ENSMUST00000077337.8
|
Irx1
|
Iroquois related homeobox 1 (Drosophila) |
| chr19_-_32061438 | 2.55 |
ENSMUST00000096119.4
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr6_-_89216237 | 2.52 |
ENSMUST00000079186.2
|
Gm839
|
predicted gene 839 |
| chr1_+_167598384 | 2.51 |
ENSMUST00000015987.3
|
Rxrg
|
retinoid X receptor gamma |
| chr17_+_64600702 | 2.51 |
ENSMUST00000086723.3
|
Man2a1
|
mannosidase 2, alpha 1 |
| chr10_+_127759721 | 2.49 |
ENSMUST00000073639.5
|
Rdh1
|
retinol dehydrogenase 1 (all trans) |
| chr2_+_109917639 | 2.43 |
ENSMUST00000046548.7
ENSMUST00000111037.2 |
Lgr4
|
leucine-rich repeat-containing G protein-coupled receptor 4 |
| chr9_-_50555170 | 2.42 |
ENSMUST00000119103.1
|
Bco2
|
beta-carotene oxygenase 2 |
| chr12_+_108334341 | 2.39 |
ENSMUST00000021684.4
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr8_-_93229517 | 2.38 |
ENSMUST00000176282.1
ENSMUST00000034173.7 |
Ces1e
|
carboxylesterase 1E |
| chr5_+_43662373 | 2.31 |
ENSMUST00000048150.8
|
Cc2d2a
|
coiled-coil and C2 domain containing 2A |
| chr2_-_35100677 | 2.29 |
ENSMUST00000045776.4
ENSMUST00000113050.3 |
AI182371
|
expressed sequence AI182371 |
| chr7_-_139582790 | 2.26 |
ENSMUST00000106095.2
|
Nkx6-2
|
NK6 homeobox 2 |
| chr2_-_58052832 | 2.21 |
ENSMUST00000090940.5
|
Ermn
|
ermin, ERM-like protein |
| chr5_+_102768771 | 2.20 |
ENSMUST00000112852.1
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr6_+_121300227 | 2.18 |
ENSMUST00000064580.7
|
Slc6a13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13 |
| chr2_+_58755177 | 2.18 |
ENSMUST00000102755.3
|
Upp2
|
uridine phosphorylase 2 |
| chr3_+_138374121 | 2.17 |
ENSMUST00000171054.1
|
Adh6-ps1
|
alcohol dehydrogenase 6 (class V), pseudogene 1 |
| chr8_-_91801948 | 2.12 |
ENSMUST00000175795.1
|
Irx3
|
Iroquois related homeobox 3 (Drosophila) |
| chr3_+_94372794 | 2.10 |
ENSMUST00000029795.3
|
Rorc
|
RAR-related orphan receptor gamma |
| chr11_-_31671727 | 2.08 |
ENSMUST00000109415.1
|
Bod1
|
biorientation of chromosomes in cell division 1 |
| chr1_-_65179058 | 2.05 |
ENSMUST00000097709.4
|
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chrX_-_75875101 | 2.04 |
ENSMUST00000114059.3
|
Pls3
|
plastin 3 (T-isoform) |
| chrX_-_8193387 | 2.03 |
ENSMUST00000143223.1
ENSMUST00000033509.8 |
Ebp
|
phenylalkylamine Ca2+ antagonist (emopamil) binding protein |
| chr4_+_136143497 | 2.03 |
ENSMUST00000008016.2
|
Id3
|
inhibitor of DNA binding 3 |
| chr5_-_88676135 | 2.03 |
ENSMUST00000078945.5
|
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr1_-_140183404 | 2.01 |
ENSMUST00000066859.6
ENSMUST00000111976.2 |
Cfh
|
complement component factor h |
| chr9_-_103219823 | 2.00 |
ENSMUST00000168142.1
|
Trf
|
transferrin |
| chr1_-_140183283 | 1.99 |
ENSMUST00000111977.1
|
Cfh
|
complement component factor h |
| chr7_-_19796789 | 1.96 |
ENSMUST00000108449.2
ENSMUST00000043822.7 |
Cblc
|
Casitas B-lineage lymphoma c |
| chr1_-_168431695 | 1.95 |
ENSMUST00000176790.1
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr10_+_34297421 | 1.94 |
ENSMUST00000047935.6
|
Tspyl4
|
TSPY-like 4 |
| chr17_-_12940317 | 1.93 |
ENSMUST00000160378.1
ENSMUST00000043923.5 |
Acat3
|
acetyl-Coenzyme A acetyltransferase 3 |
| chr11_-_70514608 | 1.92 |
ENSMUST00000021179.3
|
Vmo1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chr7_+_19489045 | 1.91 |
ENSMUST00000011407.7
ENSMUST00000137613.1 |
Exoc3l2
|
exocyst complex component 3-like 2 |
| chr14_-_36968679 | 1.90 |
ENSMUST00000067700.6
|
Ccser2
|
coiled-coil serine rich 2 |
| chr1_+_169969409 | 1.87 |
ENSMUST00000180638.1
|
3110045C21Rik
|
RIKEN cDNA 3110045C21 gene |
| chr2_-_52335134 | 1.87 |
ENSMUST00000075301.3
|
Neb
|
nebulin |
| chr2_-_150451486 | 1.86 |
ENSMUST00000109916.1
|
Zfp442
|
zinc finger protein 442 |
| chr9_-_21927515 | 1.82 |
ENSMUST00000178988.1
ENSMUST00000046831.9 |
Tmem205
|
transmembrane protein 205 |
| chr18_-_61536522 | 1.82 |
ENSMUST00000171629.1
|
Arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr7_+_44590886 | 1.80 |
ENSMUST00000107906.3
|
Kcnc3
|
potassium voltage gated channel, Shaw-related subfamily, member 3 |
| chr12_-_56613270 | 1.79 |
ENSMUST00000072631.5
|
Nkx2-9
|
NK2 homeobox 9 |
| chr11_-_86993682 | 1.79 |
ENSMUST00000018571.4
|
Ypel2
|
yippee-like 2 (Drosophila) |
| chr13_-_56548534 | 1.78 |
ENSMUST00000062806.4
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr14_+_21052574 | 1.77 |
ENSMUST00000045376.9
|
Adk
|
adenosine kinase |
| chr9_+_77921908 | 1.76 |
ENSMUST00000133757.1
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
| chr6_+_82041623 | 1.76 |
ENSMUST00000042974.8
|
Eva1a
|
eva-1 homolog A (C. elegans) |
| chr4_-_41640322 | 1.76 |
ENSMUST00000127306.1
|
Enho
|
energy homeostasis associated |
| chr16_+_45093611 | 1.72 |
ENSMUST00000099498.2
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr8_-_83955205 | 1.70 |
ENSMUST00000098595.2
|
Gm10644
|
predicted gene 10644 |
| chr8_-_3878549 | 1.67 |
ENSMUST00000011445.6
|
Cd209d
|
CD209d antigen |
| chr4_-_104876383 | 1.67 |
ENSMUST00000064873.8
ENSMUST00000106808.3 ENSMUST00000048947.8 |
C8a
|
complement component 8, alpha polypeptide |
| chr11_+_117232254 | 1.66 |
ENSMUST00000106354.2
|
Sept9
|
septin 9 |
| chr15_-_55113460 | 1.65 |
ENSMUST00000100659.2
ENSMUST00000110230.1 |
Gm9920
|
predicted gene 9920 |
| chr4_-_109156610 | 1.65 |
ENSMUST00000161363.1
|
Osbpl9
|
oxysterol binding protein-like 9 |
| chr6_+_129180613 | 1.64 |
ENSMUST00000032260.5
|
Clec2d
|
C-type lectin domain family 2, member d |
| chr9_+_21927471 | 1.59 |
ENSMUST00000170304.1
ENSMUST00000006403.6 |
Ccdc159
|
coiled-coil domain containing 159 |
| chr9_+_70207342 | 1.58 |
ENSMUST00000034745.7
|
Myo1e
|
myosin IE |
| chr17_-_47421873 | 1.52 |
ENSMUST00000073143.6
|
1700001C19Rik
|
RIKEN cDNA 1700001C19 gene |
| chr14_-_61556746 | 1.52 |
ENSMUST00000100496.4
|
Spryd7
|
SPRY domain containing 7 |
| chr6_-_23132981 | 1.52 |
ENSMUST00000031707.7
|
Aass
|
aminoadipate-semialdehyde synthase |
| chr7_-_25539845 | 1.51 |
ENSMUST00000066503.7
ENSMUST00000064862.6 |
Ceacam2
|
carcinoembryonic antigen-related cell adhesion molecule 2 |
| chr13_+_4049001 | 1.49 |
ENSMUST00000118717.2
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr6_-_72362382 | 1.47 |
ENSMUST00000114095.1
ENSMUST00000069595.6 ENSMUST00000069580.5 |
Rnf181
|
ring finger protein 181 |
| chr4_+_89137122 | 1.45 |
ENSMUST00000058030.7
|
Mtap
|
methylthioadenosine phosphorylase |
| chr7_-_137410717 | 1.45 |
ENSMUST00000120340.1
ENSMUST00000117404.1 ENSMUST00000068996.6 |
9430038I01Rik
|
RIKEN cDNA 9430038I01 gene |
| chr4_+_97777780 | 1.45 |
ENSMUST00000107062.2
ENSMUST00000052018.5 ENSMUST00000107057.1 |
Nfia
|
nuclear factor I/A |
| chr10_-_54075730 | 1.44 |
ENSMUST00000105469.1
ENSMUST00000003843.8 |
Man1a
|
mannosidase 1, alpha |
| chr11_+_83709015 | 1.44 |
ENSMUST00000001009.7
|
Wfdc18
|
WAP four-disulfide core domain 18 |
| chr13_+_63015167 | 1.43 |
ENSMUST00000021911.8
|
2010111I01Rik
|
RIKEN cDNA 2010111I01 gene |
| chrX_-_147429189 | 1.42 |
ENSMUST00000033646.2
|
Il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr2_-_104742802 | 1.42 |
ENSMUST00000028595.7
|
Depdc7
|
DEP domain containing 7 |
| chr4_+_135946447 | 1.42 |
ENSMUST00000030432.7
|
Hmgcl
|
3-hydroxy-3-methylglutaryl-Coenzyme A lyase |
| chr14_+_55559993 | 1.41 |
ENSMUST00000117236.1
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr4_+_43046014 | 1.40 |
ENSMUST00000180426.1
|
Gm26881
|
predicted gene, 26881 |
| chr14_+_123659971 | 1.39 |
ENSMUST00000049681.7
|
Itgbl1
|
integrin, beta-like 1 |
| chr17_-_46032366 | 1.39 |
ENSMUST00000071648.5
ENSMUST00000142351.2 ENSMUST00000167860.1 |
Vegfa
|
vascular endothelial growth factor A |
| chr14_-_55560340 | 1.39 |
ENSMUST00000066106.3
|
A730061H03Rik
|
RIKEN cDNA A730061H03 gene |
| chr8_-_77610597 | 1.38 |
ENSMUST00000034030.8
|
Tmem184c
|
transmembrane protein 184C |
| chr1_-_169969143 | 1.38 |
ENSMUST00000027989.6
ENSMUST00000111353.3 |
Hsd17b7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
| chr7_+_101378183 | 1.37 |
ENSMUST00000084895.5
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr17_-_36058371 | 1.37 |
ENSMUST00000113742.2
|
Gm11127
|
predicted gene 11127 |
| chr13_-_62888282 | 1.36 |
ENSMUST00000092888.4
|
Fbp1
|
fructose bisphosphatase 1 |
| chr1_+_152399824 | 1.36 |
ENSMUST00000044311.8
|
Colgalt2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr13_+_55714624 | 1.36 |
ENSMUST00000021959.9
|
Txndc15
|
thioredoxin domain containing 15 |
| chr19_+_46623387 | 1.35 |
ENSMUST00000111855.4
|
Wbp1l
|
WW domain binding protein 1 like |
| chrX_+_38600626 | 1.34 |
ENSMUST00000000365.2
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chr10_+_94036001 | 1.34 |
ENSMUST00000020208.4
|
Fgd6
|
FYVE, RhoGEF and PH domain containing 6 |
| chrX_+_142226765 | 1.34 |
ENSMUST00000112916.2
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr14_+_12189943 | 1.33 |
ENSMUST00000119888.1
|
Ptprg
|
protein tyrosine phosphatase, receptor type, G |
| chrX_+_103356464 | 1.33 |
ENSMUST00000116547.2
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr2_+_127270208 | 1.33 |
ENSMUST00000110375.2
|
Stard7
|
START domain containing 7 |
| chr8_+_45627709 | 1.33 |
ENSMUST00000134321.1
ENSMUST00000135336.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr7_-_45103747 | 1.32 |
ENSMUST00000003512.7
|
Fcgrt
|
Fc receptor, IgG, alpha chain transporter |
| chrX_+_129749740 | 1.32 |
ENSMUST00000167619.2
ENSMUST00000037854.8 |
Diap2
|
diaphanous homolog 2 (Drosophila) |
| chr14_+_55560010 | 1.32 |
ENSMUST00000147981.1
ENSMUST00000133256.1 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr4_+_116596672 | 1.32 |
ENSMUST00000051869.7
|
Ccdc17
|
coiled-coil domain containing 17 |
| chr19_+_26749726 | 1.31 |
ENSMUST00000175842.1
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_+_19957037 | 1.31 |
ENSMUST00000091309.5
ENSMUST00000108329.1 ENSMUST00000003714.6 |
Cp
|
ceruloplasmin |
| chr13_+_42301270 | 1.30 |
ENSMUST00000021796.7
|
Edn1
|
endothelin 1 |
| chr7_-_5805445 | 1.30 |
ENSMUST00000075085.6
|
Vmn1r63
|
vomeronasal 1 receptor 63 |
| chr8_-_77610668 | 1.29 |
ENSMUST00000141202.1
ENSMUST00000152168.1 |
Tmem184c
|
transmembrane protein 184C |
| chr4_+_43406435 | 1.29 |
ENSMUST00000098106.2
ENSMUST00000139198.1 |
Rusc2
|
RUN and SH3 domain containing 2 |
| chr4_+_150855064 | 1.29 |
ENSMUST00000030811.1
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr17_-_34959232 | 1.29 |
ENSMUST00000165202.1
ENSMUST00000172753.1 |
Hspa1b
|
heat shock protein 1B |
| chrX_+_129749830 | 1.28 |
ENSMUST00000113320.2
|
Diap2
|
diaphanous homolog 2 (Drosophila) |
| chr15_+_99295087 | 1.27 |
ENSMUST00000128352.1
ENSMUST00000145482.1 |
Prpf40b
|
PRP40 pre-mRNA processing factor 40 homolog B (yeast) |
| chr11_-_11898044 | 1.27 |
ENSMUST00000066237.3
|
Ddc
|
dopa decarboxylase |
| chr9_+_20581296 | 1.26 |
ENSMUST00000115557.2
|
Zfp846
|
zinc finger protein 846 |
| chr16_+_45094036 | 1.26 |
ENSMUST00000061050.5
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr4_-_154899077 | 1.26 |
ENSMUST00000030935.3
ENSMUST00000132281.1 |
Fam213b
|
family with sequence similarity 213, member B |
| chr9_+_44084944 | 1.25 |
ENSMUST00000176416.1
ENSMUST00000065461.7 |
Usp2
|
ubiquitin specific peptidase 2 |
| chr11_+_94328242 | 1.24 |
ENSMUST00000021227.5
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr8_-_41215146 | 1.23 |
ENSMUST00000034003.4
|
Fgl1
|
fibrinogen-like protein 1 |
| chr10_+_18469958 | 1.23 |
ENSMUST00000162891.1
ENSMUST00000100054.3 |
Nhsl1
|
NHS-like 1 |
| chr7_+_66079643 | 1.22 |
ENSMUST00000101801.5
|
Vimp
|
VCP-interacting membrane protein |
| chr8_-_72435043 | 1.22 |
ENSMUST00000109974.1
|
Calr3
|
calreticulin 3 |
| chr4_-_6275629 | 1.22 |
ENSMUST00000029905.1
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr11_-_11898092 | 1.22 |
ENSMUST00000178704.1
|
Ddc
|
dopa decarboxylase |
| chr2_+_177897096 | 1.22 |
ENSMUST00000108935.1
|
Gm14327
|
predicted gene 14327 |
| chr14_-_72602945 | 1.21 |
ENSMUST00000162825.1
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chr1_+_185332143 | 1.21 |
ENSMUST00000027916.6
ENSMUST00000151769.1 ENSMUST00000110965.1 |
Bpnt1
|
bisphosphate 3'-nucleotidase 1 |
| chr11_+_87592145 | 1.21 |
ENSMUST00000103179.3
ENSMUST00000092802.5 ENSMUST00000146871.1 |
Mtmr4
|
myotubularin related protein 4 |
| chr1_+_21240581 | 1.21 |
ENSMUST00000027067.8
|
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr1_+_21240597 | 1.20 |
ENSMUST00000121676.1
|
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr17_-_46890405 | 1.20 |
ENSMUST00000086675.3
|
A330017A19Rik
|
RIKEN cDNA A330017A19 gene |
| chr17_+_36121666 | 1.20 |
ENSMUST00000173128.1
|
Gm19684
|
predicted gene, 19684 |
| chr7_-_80405425 | 1.19 |
ENSMUST00000107362.3
ENSMUST00000135306.1 |
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr10_-_89506631 | 1.18 |
ENSMUST00000058126.8
ENSMUST00000105296.2 |
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr5_-_137684665 | 1.18 |
ENSMUST00000100544.4
ENSMUST00000031736.9 ENSMUST00000151839.1 |
Agfg2
|
ArfGAP with FG repeats 2 |
| chr8_-_3748941 | 1.17 |
ENSMUST00000012847.1
|
Cd209a
|
CD209a antigen |
| chr3_-_107943705 | 1.17 |
ENSMUST00000106680.1
ENSMUST00000106684.1 ENSMUST00000106685.2 |
Gstm6
|
glutathione S-transferase, mu 6 |
| chr17_-_91092715 | 1.15 |
ENSMUST00000160800.2
ENSMUST00000159778.1 ENSMUST00000160844.3 |
Nrxn1
|
neurexin I |
| chr13_+_16011851 | 1.15 |
ENSMUST00000042603.6
|
Inhba
|
inhibin beta-A |
| chr10_-_75780954 | 1.15 |
ENSMUST00000173512.1
ENSMUST00000173537.1 |
Gm20441
Gstt3
|
predicted gene 20441 glutathione S-transferase, theta 3 |
| chr2_+_143915273 | 1.15 |
ENSMUST00000103172.3
|
Dstn
|
destrin |
| chr13_+_9276477 | 1.15 |
ENSMUST00000174552.1
|
Dip2c
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr7_-_140116395 | 1.14 |
ENSMUST00000026538.6
|
Echs1
|
enoyl Coenzyme A hydratase, short chain, 1, mitochondrial |
| chr1_+_88138364 | 1.14 |
ENSMUST00000014263.4
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr7_-_25754701 | 1.13 |
ENSMUST00000108401.1
ENSMUST00000043765.7 |
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr2_-_64975762 | 1.13 |
ENSMUST00000156765.1
|
Grb14
|
growth factor receptor bound protein 14 |
| chr4_+_97777606 | 1.13 |
ENSMUST00000075448.6
ENSMUST00000092532.6 |
Nfia
|
nuclear factor I/A |
| chr11_+_70540064 | 1.13 |
ENSMUST00000157075.1
|
Pld2
|
phospholipase D2 |
| chr11_+_95842283 | 1.12 |
ENSMUST00000107714.2
ENSMUST00000107711.1 |
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr3_-_97297778 | 1.12 |
ENSMUST00000181368.1
|
Gm17608
|
predicted gene, 17608 |
| chr11_+_71019593 | 1.12 |
ENSMUST00000133413.1
ENSMUST00000164220.1 ENSMUST00000048807.5 |
Mis12
|
MIS12 homolog (yeast) |
| chr11_-_115187827 | 1.11 |
ENSMUST00000103041.1
|
Nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr3_-_148989316 | 1.10 |
ENSMUST00000098518.2
|
Lphn2
|
latrophilin 2 |
| chr7_+_4237699 | 1.10 |
ENSMUST00000117550.1
|
Lilra5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| chr5_-_66151903 | 1.09 |
ENSMUST00000167950.1
|
Rbm47
|
RNA binding motif protein 47 |
| chr2_-_26092149 | 1.09 |
ENSMUST00000114159.2
|
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chr7_-_101933815 | 1.09 |
ENSMUST00000106963.1
ENSMUST00000106966.1 |
Lrrc51
|
leucine rich repeat containing 51 |
| chr19_+_11912389 | 1.09 |
ENSMUST00000061618.7
|
Patl1
|
protein associated with topoisomerase II homolog 1 (yeast) |
| chr14_-_45530118 | 1.08 |
ENSMUST00000045905.6
|
Fermt2
|
fermitin family homolog 2 (Drosophila) |
| chr6_-_87851011 | 1.08 |
ENSMUST00000113617.1
|
Cnbp
|
cellular nucleic acid binding protein |
| chr11_-_23770953 | 1.08 |
ENSMUST00000102864.3
|
Rel
|
reticuloendotheliosis oncogene |
| chr19_+_44989073 | 1.07 |
ENSMUST00000026225.8
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gfi1_Gfi1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.6 | GO:0032385 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 1.5 | 4.6 | GO:0070949 | neutrophil mediated killing of fungus(GO:0070947) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 1.1 | 3.4 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 1.0 | 3.0 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 1.0 | 4.0 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.9 | 2.7 | GO:0035622 | intrahepatic bile duct development(GO:0035622) renal vesicle induction(GO:0072034) |
| 0.8 | 2.4 | GO:0060995 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.8 | 2.4 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 0.8 | 3.2 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.8 | 3.2 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.8 | 12.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.7 | 2.9 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.7 | 2.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.7 | 4.7 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.6 | 3.9 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.6 | 1.9 | GO:0038189 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.6 | 3.8 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.5 | 1.5 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.5 | 1.5 | GO:0070342 | brown fat cell proliferation(GO:0070342) regulation of brown fat cell proliferation(GO:0070347) |
| 0.4 | 1.2 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 0.4 | 2.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.4 | 1.5 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.4 | 1.1 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.4 | 0.7 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.4 | 1.1 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.4 | 2.5 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.4 | 1.4 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.3 | 1.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.3 | 1.0 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.3 | 1.4 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.3 | 0.3 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 0.3 | 2.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.3 | 2.3 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.3 | 4.5 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.3 | 1.6 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.3 | 5.8 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.3 | 1.2 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.3 | 1.2 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.3 | 1.2 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.3 | 3.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.3 | 1.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.3 | 2.0 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.3 | 2.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.3 | 1.3 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.3 | 9.9 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.2 | 1.0 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) mesenchymal stem cell proliferation(GO:0097168) |
| 0.2 | 2.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 3.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.2 | 3.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 0.9 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.2 | 1.5 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.2 | 0.6 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.2 | 0.6 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.2 | 0.6 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.2 | 0.6 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.2 | 2.6 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 3.4 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.2 | 2.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 1.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 0.9 | GO:0032349 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.2 | 0.6 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.2 | 0.7 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.2 | 1.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.2 | 0.6 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.2 | 0.2 | GO:2000137 | negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 0.2 | 0.7 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.2 | 1.6 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.2 | 1.4 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.2 | 0.7 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.2 | 1.7 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 0.8 | GO:0061744 | motor behavior(GO:0061744) |
| 0.2 | 1.9 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.2 | 1.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.2 | 2.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.2 | 0.9 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.2 | 0.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.4 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 1.8 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.3 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 1.0 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 3.0 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.7 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.8 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 | 4.1 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 0.8 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.9 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 0.7 | GO:0098961 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.1 | 0.7 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 | 1.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.7 | GO:0072423 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.1 | 0.9 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 1.0 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 1.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.4 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.4 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.1 | 3.1 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 0.3 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 0.5 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 0.4 | GO:0003420 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.1 | 0.4 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 1.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 1.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 2.3 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 1.1 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.3 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.4 | GO:1901146 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.1 | 5.0 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 2.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 2.5 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.1 | 0.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 1.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 1.0 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 2.0 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 0.7 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 1.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.3 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.6 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.1 | 1.0 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 0.1 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) |
| 0.1 | 0.2 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.1 | 0.9 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.3 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 | 0.3 | GO:0051661 | maintenance of centrosome location(GO:0051661) maintenance of Golgi location(GO:0051684) |
| 0.1 | 2.9 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 0.8 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 1.0 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 0.9 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 1.3 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 1.7 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.8 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 0.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.7 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.4 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.1 | 0.9 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.7 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 1.0 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.1 | 0.2 | GO:0043519 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.1 | 14.3 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 0.1 | GO:0072431 | signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.1 | 1.2 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.1 | 0.5 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 0.6 | GO:0048733 | histone H3-K36 methylation(GO:0010452) sebaceous gland development(GO:0048733) |
| 0.1 | 0.7 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 0.6 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 10.3 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 1.8 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.2 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 0.3 | GO:0051095 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 3.1 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.5 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.3 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 1.3 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 3.9 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.6 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.7 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 0.4 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.7 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 2.3 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.4 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.3 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.5 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.3 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 1.1 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 2.0 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:2000503 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.8 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.2 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.2 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.2 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.9 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 1.2 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.3 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.1 | GO:2000412 | tolerance induction to self antigen(GO:0002513) positive regulation of thymocyte migration(GO:2000412) |
| 0.0 | 0.5 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 1.5 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.6 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 0.1 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 1.4 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.0 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) development of secondary male sexual characteristics(GO:0046544) |
| 0.0 | 0.1 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) short-chain fatty acid metabolic process(GO:0046459) |
| 0.0 | 1.9 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 1.6 | GO:0001656 | metanephros development(GO:0001656) |
| 0.0 | 0.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 1.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.2 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.6 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 | 0.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.4 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.6 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.5 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 1.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 2.2 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 1.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.0 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 0.0 | 1.6 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 1.9 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.0 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.0 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.1 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.8 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.2 | GO:0042670 | amacrine cell differentiation(GO:0035881) retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.4 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.8 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.4 | GO:0071353 | response to interleukin-4(GO:0070670) cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.7 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.4 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 2.8 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 1.9 | GO:0009166 | nucleotide catabolic process(GO:0009166) |
| 0.0 | 0.2 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.0 | 1.2 | GO:0030193 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 | 2.6 | GO:0006665 | sphingolipid metabolic process(GO:0006665) |
| 0.0 | 0.6 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.4 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 2.2 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.7 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.0 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.4 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 1.2 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.0 | GO:0071873 | epinephrine metabolic process(GO:0042414) response to norepinephrine(GO:0071873) |
| 0.0 | 0.4 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 1.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.1 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.3 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 1.3 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.6 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.5 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.2 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 1.2 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.6 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.4 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.5 | 4.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.5 | 1.9 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.4 | 1.8 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.4 | 1.1 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.3 | 3.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 1.9 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.3 | 4.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.3 | 11.0 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.3 | 2.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.3 | 1.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.2 | 3.0 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.2 | 2.2 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 1.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 0.6 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.2 | 0.4 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.2 | 0.9 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.2 | 0.6 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.2 | 0.9 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.2 | 0.7 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 3.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 1.3 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 0.5 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.6 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.8 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 4.0 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 1.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 1.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 2.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 0.3 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.1 | 1.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.8 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 2.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 3.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.8 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 1.7 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 0.3 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.1 | 0.7 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.1 | 1.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.6 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 0.7 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 1.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 3.5 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.1 | 1.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 1.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 1.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 3.0 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.6 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.6 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.5 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.8 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 1.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 2.3 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 1.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 1.5 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 6.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.0 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 1.9 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 1.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.0 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 4.2 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 1.2 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.4 | GO:0032994 | protein-lipid complex(GO:0032994) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 1.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 5.6 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 3.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 6.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 3.8 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 2.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 2.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 2.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.4 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 2.8 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 2.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0097228 | sperm principal piece(GO:0097228) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 2.0 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 1.7 | 6.6 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 1.3 | 4.0 | GO:0004024 | NADPH:quinone reductase activity(GO:0003960) alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 1.3 | 3.8 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 1.1 | 3.4 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 1.0 | 9.8 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 1.0 | 4.8 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.8 | 3.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.8 | 3.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.7 | 2.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.7 | 3.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.6 | 1.9 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.6 | 2.5 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.6 | 4.0 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.5 | 7.5 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.5 | 3.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.5 | 1.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.5 | 1.4 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.4 | 2.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.4 | 2.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.4 | 2.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.4 | 4.0 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.4 | 4.6 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.4 | 1.4 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.3 | 1.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.3 | 1.9 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.3 | 1.2 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.3 | 5.7 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.3 | 0.9 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.3 | 2.0 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.3 | 1.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.3 | 2.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 0.7 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.2 | 1.5 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.2 | 1.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.2 | 8.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 1.6 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.2 | 0.7 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.2 | 0.8 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.2 | 6.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.2 | 0.6 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 1.7 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.2 | 1.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 2.9 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 0.5 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.2 | 0.7 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.2 | 1.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 0.8 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.2 | 2.5 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 1.8 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.6 | GO:0031711 | tripeptidyl-peptidase activity(GO:0008240) bradykinin receptor binding(GO:0031711) metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.7 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 2.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 1.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.8 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.1 | 2.8 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 1.4 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.1 | 1.0 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.1 | 0.6 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.8 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 1.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.4 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 2.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.9 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 2.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.6 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 1.6 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 0.4 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.1 | 1.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 5.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 4.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 3.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 12.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 1.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 3.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.7 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 0.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 1.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.6 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 0.5 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 1.5 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 1.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.3 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 1.9 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 1.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.4 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 | 3.4 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.3 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.3 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.7 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 2.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.0 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.6 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.4 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.4 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 1.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 1.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.5 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.2 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 1.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 1.0 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.5 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 2.1 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 1.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 2.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.4 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 1.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 1.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 5.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.5 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.5 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 1.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.6 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 2.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.5 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.0 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 1.1 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 10.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 4.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 0.8 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 5.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 3.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 2.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 5.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 2.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 1.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 0.7 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 2.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 0.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.4 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 2.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 5.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.7 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.7 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.7 | 5.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.5 | 2.0 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.4 | 14.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.4 | 10.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.3 | 4.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 3.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 2.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 3.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 3.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 6.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 0.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 2.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 12.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.3 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 2.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 3.9 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.1 | 1.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 3.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 1.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 5.0 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 1.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 3.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 5.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 1.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 2.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 0.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.0 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 0.7 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 1.4 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 1.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 6.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.6 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 1.3 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |