Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
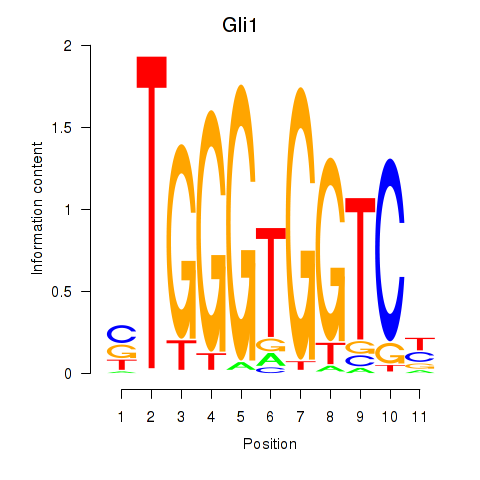
Results for Gli1
Z-value: 0.55
Transcription factors associated with Gli1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gli1
|
ENSMUSG00000025407.6 | GLI-Kruppel family member GLI1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gli1 | mm10_v2_chr10_-_127341583_127341614 | 0.02 | 9.1e-01 | Click! |
Activity profile of Gli1 motif
Sorted Z-values of Gli1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_24370255 | 1.45 |
ENSMUST00000171904.1
|
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr11_+_115899943 | 1.41 |
ENSMUST00000152171.1
|
Smim5
|
small integral membrane protein 5 |
| chr4_+_154869585 | 1.35 |
ENSMUST00000079269.7
ENSMUST00000163732.1 ENSMUST00000080559.6 |
Mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr1_-_133801031 | 1.30 |
ENSMUST00000143567.1
|
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr6_-_122609964 | 1.22 |
ENSMUST00000032211.4
|
Gdf3
|
growth differentiation factor 3 |
| chr11_-_102365111 | 1.16 |
ENSMUST00000006749.9
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr18_+_61687911 | 1.02 |
ENSMUST00000025471.2
|
Il17b
|
interleukin 17B |
| chr17_-_45733843 | 1.00 |
ENSMUST00000178179.1
|
1600014C23Rik
|
RIKEN cDNA 1600014C23 gene |
| chr17_-_24209377 | 1.00 |
ENSMUST00000024931.4
|
Ntn3
|
netrin 3 |
| chr6_-_52218686 | 0.99 |
ENSMUST00000134367.2
|
Hoxa7
|
homeobox A7 |
| chr4_-_133874682 | 0.85 |
ENSMUST00000168974.2
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr8_+_121127827 | 0.84 |
ENSMUST00000181609.1
|
Foxl1
|
forkhead box L1 |
| chrX_-_59568068 | 0.83 |
ENSMUST00000119833.1
ENSMUST00000131319.1 |
Fgf13
|
fibroblast growth factor 13 |
| chr13_+_6548154 | 0.83 |
ENSMUST00000021611.8
|
Pitrm1
|
pitrilysin metallepetidase 1 |
| chrX_+_159303266 | 0.83 |
ENSMUST00000112491.1
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr4_+_45184815 | 0.80 |
ENSMUST00000134280.1
ENSMUST00000044773.5 |
Frmpd1
|
FERM and PDZ domain containing 1 |
| chr2_-_152830615 | 0.80 |
ENSMUST00000146380.1
ENSMUST00000134902.1 ENSMUST00000134357.1 ENSMUST00000109820.3 |
Bcl2l1
|
BCL2-like 1 |
| chr1_+_129273344 | 0.80 |
ENSMUST00000073527.6
ENSMUST00000040311.7 |
Thsd7b
|
thrombospondin, type I, domain containing 7B |
| chr2_+_119351222 | 0.78 |
ENSMUST00000028780.3
|
Chac1
|
ChaC, cation transport regulator 1 |
| chr11_+_72999069 | 0.78 |
ENSMUST00000021141.7
|
P2rx1
|
purinergic receptor P2X, ligand-gated ion channel, 1 |
| chr15_-_103310425 | 0.75 |
ENSMUST00000079824.4
|
Gpr84
|
G protein-coupled receptor 84 |
| chr2_+_118111876 | 0.73 |
ENSMUST00000039559.8
|
Thbs1
|
thrombospondin 1 |
| chr13_-_117025505 | 0.69 |
ENSMUST00000022239.6
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr8_+_70501116 | 0.68 |
ENSMUST00000127983.1
|
Crlf1
|
cytokine receptor-like factor 1 |
| chr3_+_95929246 | 0.67 |
ENSMUST00000165307.1
ENSMUST00000015893.6 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr13_+_44729535 | 0.65 |
ENSMUST00000174068.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chrX_-_167382747 | 0.65 |
ENSMUST00000026839.4
|
Prps2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr7_-_118443549 | 0.64 |
ENSMUST00000081574.4
|
Syt17
|
synaptotagmin XVII |
| chr2_+_29890534 | 0.64 |
ENSMUST00000113764.3
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr3_+_95929325 | 0.63 |
ENSMUST00000171368.1
ENSMUST00000168106.1 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chrX_+_140956892 | 0.63 |
ENSMUST00000112971.1
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr13_+_109685994 | 0.61 |
ENSMUST00000074103.5
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr11_+_96282648 | 0.61 |
ENSMUST00000168043.1
|
Hoxb8
|
homeobox B8 |
| chr15_-_56694525 | 0.61 |
ENSMUST00000050544.7
|
Has2
|
hyaluronan synthase 2 |
| chr11_+_9191934 | 0.60 |
ENSMUST00000042740.6
|
Abca13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr2_-_32083783 | 0.59 |
ENSMUST00000056406.6
|
Fam78a
|
family with sequence similarity 78, member A |
| chr11_+_77009265 | 0.59 |
ENSMUST00000129572.2
|
Slc6a4
|
solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 |
| chr1_-_167466780 | 0.59 |
ENSMUST00000036643.4
|
Lrrc52
|
leucine rich repeat containing 52 |
| chr2_+_29889785 | 0.58 |
ENSMUST00000113763.1
ENSMUST00000113757.1 ENSMUST00000113756.1 ENSMUST00000133233.1 ENSMUST00000113759.2 ENSMUST00000113755.1 ENSMUST00000137558.1 ENSMUST00000046571.7 |
Odf2
|
outer dense fiber of sperm tails 2 |
| chr11_+_73240310 | 0.57 |
ENSMUST00000138853.1
|
Trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr6_-_41636389 | 0.56 |
ENSMUST00000031902.5
|
Trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr2_-_152831112 | 0.56 |
ENSMUST00000128172.1
|
Bcl2l1
|
BCL2-like 1 |
| chrX_+_103321398 | 0.54 |
ENSMUST00000033689.2
|
Cdx4
|
caudal type homeobox 4 |
| chr2_+_29890096 | 0.53 |
ENSMUST00000113762.1
ENSMUST00000113765.1 |
Odf2
|
outer dense fiber of sperm tails 2 |
| chrX_-_59567348 | 0.53 |
ENSMUST00000124402.1
|
Fgf13
|
fibroblast growth factor 13 |
| chr6_+_113531675 | 0.52 |
ENSMUST00000036340.5
ENSMUST00000101051.2 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr4_-_148130678 | 0.50 |
ENSMUST00000030862.4
|
Draxin
|
dorsal inhibitory axon guidance protein |
| chr2_+_29890063 | 0.49 |
ENSMUST00000028128.6
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr8_-_64733534 | 0.49 |
ENSMUST00000141021.1
|
Sc4mol
|
sterol-C4-methyl oxidase-like |
| chr18_-_70141568 | 0.46 |
ENSMUST00000121693.1
|
Rab27b
|
RAB27b, member RAS oncogene family |
| chr2_-_59948155 | 0.45 |
ENSMUST00000153136.1
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr2_+_29889720 | 0.45 |
ENSMUST00000113767.1
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr11_-_34833631 | 0.45 |
ENSMUST00000093191.2
|
Spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr7_-_44227421 | 0.44 |
ENSMUST00000072123.4
|
Gm10109
|
predicted gene 10109 |
| chr15_-_95528702 | 0.44 |
ENSMUST00000166170.1
|
Nell2
|
NEL-like 2 |
| chr18_-_35662180 | 0.43 |
ENSMUST00000025209.4
ENSMUST00000096573.2 |
Spata24
|
spermatogenesis associated 24 |
| chr16_-_91011029 | 0.43 |
ENSMUST00000130813.1
|
Synj1
|
synaptojanin 1 |
| chr16_-_91011093 | 0.42 |
ENSMUST00000170853.1
ENSMUST00000118390.2 |
Synj1
|
synaptojanin 1 |
| chr7_-_6331235 | 0.41 |
ENSMUST00000127658.1
ENSMUST00000062765.7 |
Zfp583
|
zinc finger protein 583 |
| chr4_+_118428078 | 0.40 |
ENSMUST00000006557.6
ENSMUST00000167636.1 ENSMUST00000102673.4 |
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chr5_-_110779937 | 0.40 |
ENSMUST00000112426.1
|
Pus1
|
pseudouridine synthase 1 |
| chr7_+_28982832 | 0.39 |
ENSMUST00000085835.6
|
Map4k1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr16_-_56024628 | 0.38 |
ENSMUST00000119981.1
ENSMUST00000096021.3 |
Pcnp
|
PEST proteolytic signal containing nuclear protein |
| chr7_+_27499315 | 0.38 |
ENSMUST00000098644.2
ENSMUST00000108355.1 |
Prx
|
periaxin |
| chr6_-_148944750 | 0.37 |
ENSMUST00000111562.1
ENSMUST00000081956.5 |
Fam60a
|
family with sequence similarity 60, member A |
| chr6_-_52246214 | 0.35 |
ENSMUST00000048026.8
|
Hoxa11
|
homeobox A11 |
| chr17_+_86963279 | 0.35 |
ENSMUST00000139344.1
|
Rhoq
|
ras homolog gene family, member Q |
| chr11_+_20543307 | 0.34 |
ENSMUST00000093292.4
|
Sertad2
|
SERTA domain containing 2 |
| chr9_-_82975475 | 0.33 |
ENSMUST00000034787.5
|
Phip
|
pleckstrin homology domain interacting protein |
| chr5_-_115300912 | 0.33 |
ENSMUST00000112090.1
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr7_-_24760311 | 0.32 |
ENSMUST00000063956.5
|
Cd177
|
CD177 antigen |
| chrX_+_101299143 | 0.32 |
ENSMUST00000118111.1
ENSMUST00000130555.1 ENSMUST00000151528.1 |
Nlgn3
|
neuroligin 3 |
| chrX_+_137049586 | 0.32 |
ENSMUST00000047852.7
|
Fam199x
|
family with sequence similarity 199, X-linked |
| chr5_-_115300957 | 0.31 |
ENSMUST00000009157.3
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr8_-_120177440 | 0.31 |
ENSMUST00000048786.6
|
Fam92b
|
family with sequence similarity 92, member B |
| chr9_+_55149364 | 0.30 |
ENSMUST00000121677.1
|
Ube2q2
|
ubiquitin-conjugating enzyme E2Q (putative) 2 |
| chr11_-_45955183 | 0.30 |
ENSMUST00000109254.1
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr9_+_118506226 | 0.30 |
ENSMUST00000084820.4
|
Golga4
|
golgi autoantigen, golgin subfamily a, 4 |
| chr15_-_95528228 | 0.29 |
ENSMUST00000075275.2
|
Nell2
|
NEL-like 2 |
| chr15_-_100669496 | 0.29 |
ENSMUST00000182814.1
ENSMUST00000182068.1 |
Bin2
|
bridging integrator 2 |
| chr13_+_35741313 | 0.29 |
ENSMUST00000163595.2
|
Cdyl
|
chromodomain protein, Y chromosome-like |
| chr11_+_51619731 | 0.29 |
ENSMUST00000127405.1
|
Nhp2
|
NHP2 ribonucleoprotein |
| chr2_+_106693185 | 0.29 |
ENSMUST00000111063.1
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr6_+_67115671 | 0.29 |
ENSMUST00000181304.1
|
A430010J10Rik
|
RIKEN cDNA A430010J10 gene |
| chr17_-_25727364 | 0.28 |
ENSMUST00000170070.1
ENSMUST00000048054.7 |
Chtf18
|
CTF18, chromosome transmission fidelity factor 18 |
| chr6_+_28423539 | 0.28 |
ENSMUST00000020717.5
ENSMUST00000169841.1 |
Arf5
|
ADP-ribosylation factor 5 |
| chr19_+_11404735 | 0.28 |
ENSMUST00000153546.1
|
Ms4a4c
|
membrane-spanning 4-domains, subfamily A, member 4C |
| chr6_+_30738044 | 0.28 |
ENSMUST00000128398.1
ENSMUST00000163949.2 ENSMUST00000124665.1 |
Mest
|
mesoderm specific transcript |
| chr15_-_100669512 | 0.28 |
ENSMUST00000182574.1
ENSMUST00000182775.1 |
Bin2
|
bridging integrator 2 |
| chr2_+_105127200 | 0.27 |
ENSMUST00000139585.1
|
Wt1
|
Wilms tumor 1 homolog |
| chr3_-_92485886 | 0.27 |
ENSMUST00000054599.7
|
Sprr1a
|
small proline-rich protein 1A |
| chr11_+_60479915 | 0.27 |
ENSMUST00000126522.1
|
Myo15
|
myosin XV |
| chr8_+_13757663 | 0.27 |
ENSMUST00000043962.8
|
Cdc16
|
CDC16 cell division cycle 16 |
| chr5_+_5573952 | 0.26 |
ENSMUST00000101627.2
|
Gm8773
|
predicted gene 8773 |
| chr13_+_81783220 | 0.26 |
ENSMUST00000022009.8
|
Cetn3
|
centrin 3 |
| chr14_+_25607797 | 0.26 |
ENSMUST00000160229.1
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr2_+_157560078 | 0.26 |
ENSMUST00000153739.2
ENSMUST00000173595.1 ENSMUST00000109526.1 ENSMUST00000173839.1 ENSMUST00000173041.1 ENSMUST00000173793.1 ENSMUST00000172487.1 ENSMUST00000088484.5 |
Nnat
|
neuronatin |
| chr15_-_78544345 | 0.26 |
ENSMUST00000053239.2
|
Sstr3
|
somatostatin receptor 3 |
| chr13_+_118714678 | 0.26 |
ENSMUST00000022246.8
|
Fgf10
|
fibroblast growth factor 10 |
| chrX_+_155262443 | 0.25 |
ENSMUST00000026324.9
|
Acot9
|
acyl-CoA thioesterase 9 |
| chr15_-_51991679 | 0.25 |
ENSMUST00000022927.9
|
Rad21
|
RAD21 homolog (S. pombe) |
| chr17_-_89910449 | 0.24 |
ENSMUST00000086423.4
|
Gm10184
|
predicted pseudogene 10184 |
| chr11_+_62847111 | 0.23 |
ENSMUST00000150989.1
ENSMUST00000176577.1 |
Fbxw10
|
F-box and WD-40 domain protein 10 |
| chr11_+_62847062 | 0.23 |
ENSMUST00000036085.4
|
Fbxw10
|
F-box and WD-40 domain protein 10 |
| chr1_-_64121456 | 0.23 |
ENSMUST00000142009.1
ENSMUST00000114086.1 |
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr7_-_102494773 | 0.23 |
ENSMUST00000058750.3
|
Olfr545
|
olfactory receptor 545 |
| chr4_+_12089373 | 0.23 |
ENSMUST00000095143.2
ENSMUST00000063839.5 |
Rbm12b2
|
RNA binding motif protein 12 B2 |
| chr11_+_23306910 | 0.23 |
ENSMUST00000137823.1
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr1_+_133350510 | 0.22 |
ENSMUST00000094556.2
|
Ren1
|
renin 1 structural |
| chr2_-_103797617 | 0.22 |
ENSMUST00000028607.6
|
Caprin1
|
cell cycle associated protein 1 |
| chr11_-_83462961 | 0.22 |
ENSMUST00000021020.6
ENSMUST00000119346.1 ENSMUST00000103209.3 ENSMUST00000108137.2 |
Mmp28
|
matrix metallopeptidase 28 (epilysin) |
| chr17_-_32403526 | 0.22 |
ENSMUST00000137458.1
|
Rasal3
|
RAS protein activator like 3 |
| chr2_+_155013531 | 0.21 |
ENSMUST00000029123.2
|
a
|
nonagouti |
| chr11_+_23306884 | 0.21 |
ENSMUST00000180046.1
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr4_+_114821722 | 0.21 |
ENSMUST00000137570.1
|
Gm12830
|
predicted gene 12830 |
| chr11_-_45955465 | 0.21 |
ENSMUST00000011398.6
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr4_+_139193996 | 0.20 |
ENSMUST00000030518.9
|
Capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr17_-_32403551 | 0.20 |
ENSMUST00000135618.1
ENSMUST00000063824.7 |
Rasal3
|
RAS protein activator like 3 |
| chr10_+_80249106 | 0.20 |
ENSMUST00000105364.1
|
Ndufs7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7 |
| chrX_-_140956675 | 0.20 |
ENSMUST00000033805.8
ENSMUST00000112978.1 |
Psmd10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr10_-_63421739 | 0.19 |
ENSMUST00000054760.4
|
Gm7075
|
predicted gene 7075 |
| chr4_-_117682233 | 0.19 |
ENSMUST00000102687.3
|
Dmap1
|
DNA methyltransferase 1-associated protein 1 |
| chr12_-_103338314 | 0.18 |
ENSMUST00000149431.1
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr13_+_5861489 | 0.18 |
ENSMUST00000000080.6
|
Klf6
|
Kruppel-like factor 6 |
| chr4_-_40948196 | 0.18 |
ENSMUST00000030125.4
ENSMUST00000108089.1 |
Bag1
|
BCL2-associated athanogene 1 |
| chr3_+_45378396 | 0.18 |
ENSMUST00000166126.1
ENSMUST00000170695.1 ENSMUST00000171554.1 |
Pcdh10
|
protocadherin 10 |
| chr9_-_54661666 | 0.18 |
ENSMUST00000128624.1
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chrX_+_101299207 | 0.18 |
ENSMUST00000065858.2
|
Nlgn3
|
neuroligin 3 |
| chr2_+_119034783 | 0.17 |
ENSMUST00000028796.1
|
Rpusd2
|
RNA pseudouridylate synthase domain containing 2 |
| chr7_+_80186835 | 0.17 |
ENSMUST00000107383.1
ENSMUST00000032754.7 |
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr10_+_79777261 | 0.17 |
ENSMUST00000020575.4
|
Fstl3
|
follistatin-like 3 |
| chr11_-_42000284 | 0.17 |
ENSMUST00000109292.2
ENSMUST00000109290.1 |
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr1_-_192834719 | 0.16 |
ENSMUST00000057543.2
|
A730013G03Rik
|
RIKEN cDNA A730013G03 gene |
| chr8_-_105966038 | 0.16 |
ENSMUST00000116429.2
ENSMUST00000034370.9 |
Slc12a4
|
solute carrier family 12, member 4 |
| chr11_-_98149551 | 0.15 |
ENSMUST00000103143.3
|
Fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr7_-_140950236 | 0.15 |
ENSMUST00000026562.4
|
Ifitm5
|
interferon induced transmembrane protein 5 |
| chr11_-_42000532 | 0.15 |
ENSMUST00000070735.3
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr2_-_164071089 | 0.14 |
ENSMUST00000018466.3
|
Tomm34
|
translocase of outer mitochondrial membrane 34 |
| chr11_+_97840780 | 0.14 |
ENSMUST00000054783.4
|
B230217C12Rik
|
RIKEN cDNA B230217C12 gene |
| chr2_-_93046053 | 0.14 |
ENSMUST00000111272.1
ENSMUST00000178666.1 ENSMUST00000147339.1 |
Prdm11
|
PR domain containing 11 |
| chr4_-_154899077 | 0.14 |
ENSMUST00000030935.3
ENSMUST00000132281.1 |
Fam213b
|
family with sequence similarity 213, member B |
| chr10_+_69534208 | 0.14 |
ENSMUST00000182439.1
ENSMUST00000092434.5 ENSMUST00000092432.5 ENSMUST00000092431.5 ENSMUST00000054167.8 ENSMUST00000047061.6 |
Ank3
|
ankyrin 3, epithelial |
| chr5_-_122900267 | 0.13 |
ENSMUST00000031435.7
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr11_-_61494173 | 0.13 |
ENSMUST00000101085.2
ENSMUST00000079080.6 ENSMUST00000108714.1 |
Mapk7
|
mitogen-activated protein kinase 7 |
| chr14_-_54409469 | 0.13 |
ENSMUST00000000984.4
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr10_+_22645011 | 0.13 |
ENSMUST00000042261.4
|
Slc2a12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr2_+_155013430 | 0.12 |
ENSMUST00000148402.1
|
a
|
nonagouti |
| chr2_+_181497223 | 0.12 |
ENSMUST00000108799.3
|
Tpd52l2
|
tumor protein D52-like 2 |
| chr4_-_41275091 | 0.11 |
ENSMUST00000030143.6
ENSMUST00000108068.1 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr1_-_64121389 | 0.11 |
ENSMUST00000055001.3
|
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr12_-_81333129 | 0.11 |
ENSMUST00000085238.6
ENSMUST00000182208.1 |
Slc8a3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr8_+_54954728 | 0.11 |
ENSMUST00000033915.7
|
Gpm6a
|
glycoprotein m6a |
| chr4_+_108847827 | 0.10 |
ENSMUST00000102738.2
|
Kti12
|
KTI12 homolog, chromatin associated (S. cerevisiae) |
| chr11_+_85311232 | 0.10 |
ENSMUST00000020835.9
|
Ppm1d
|
protein phosphatase 1D magnesium-dependent, delta isoform |
| chr18_-_10610048 | 0.10 |
ENSMUST00000115864.1
|
Esco1
|
establishment of cohesion 1 homolog 1 (S. cerevisiae) |
| chr7_-_80340152 | 0.10 |
ENSMUST00000133728.1
|
Unc45a
|
unc-45 homolog A (C. elegans) |
| chr6_-_135310347 | 0.10 |
ENSMUST00000050471.2
|
Pbp2
|
phosphatidylethanolamine binding protein 2 |
| chr12_+_44328882 | 0.10 |
ENSMUST00000020939.8
ENSMUST00000110748.2 |
Nrcam
|
neuron-glia-CAM-related cell adhesion molecule |
| chr2_+_181497165 | 0.10 |
ENSMUST00000149163.1
ENSMUST00000000844.8 ENSMUST00000184849.1 ENSMUST00000108800.1 ENSMUST00000069712.2 |
Tpd52l2
|
tumor protein D52-like 2 |
| chr14_-_41008256 | 0.09 |
ENSMUST00000136661.1
|
Fam213a
|
family with sequence similarity 213, member A |
| chr2_+_153108468 | 0.09 |
ENSMUST00000109799.1
ENSMUST00000003370.7 |
Hck
|
hemopoietic cell kinase |
| chr16_+_92498122 | 0.09 |
ENSMUST00000023670.3
|
Clic6
|
chloride intracellular channel 6 |
| chr15_-_101940255 | 0.09 |
ENSMUST00000023799.7
|
Krt79
|
keratin 79 |
| chr4_-_147848358 | 0.09 |
ENSMUST00000105718.1
ENSMUST00000135798.1 |
Zfp933
|
zinc finger protein 933 |
| chr12_-_112673944 | 0.09 |
ENSMUST00000130342.1
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chr18_-_10610346 | 0.09 |
ENSMUST00000025142.5
|
Esco1
|
establishment of cohesion 1 homolog 1 (S. cerevisiae) |
| chr7_-_142899985 | 0.09 |
ENSMUST00000000219.3
|
Th
|
tyrosine hydroxylase |
| chr17_+_23898717 | 0.09 |
ENSMUST00000088598.5
|
Dcpp2
|
demilune cell and parotid protein 2 |
| chr4_+_40948401 | 0.09 |
ENSMUST00000030128.5
|
Chmp5
|
charged multivesicular body protein 5 |
| chr10_+_69533803 | 0.08 |
ENSMUST00000182155.1
ENSMUST00000183169.1 ENSMUST00000183148.1 |
Ank3
|
ankyrin 3, epithelial |
| chr5_-_122989086 | 0.08 |
ENSMUST00000046073.9
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr6_-_52012476 | 0.08 |
ENSMUST00000078214.5
|
Skap2
|
src family associated phosphoprotein 2 |
| chr2_+_129800451 | 0.08 |
ENSMUST00000165413.2
ENSMUST00000166282.2 |
Stk35
|
serine/threonine kinase 35 |
| chr18_-_10610124 | 0.08 |
ENSMUST00000097670.3
|
Esco1
|
establishment of cohesion 1 homolog 1 (S. cerevisiae) |
| chr11_-_78245942 | 0.08 |
ENSMUST00000002121.4
|
Supt6
|
suppressor of Ty 6 |
| chr11_-_69544556 | 0.08 |
ENSMUST00000035539.5
|
Dnah2
|
dynein, axonemal, heavy chain 2 |
| chr10_-_64090241 | 0.07 |
ENSMUST00000133588.1
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr13_-_58385190 | 0.07 |
ENSMUST00000022032.5
|
2210016F16Rik
|
RIKEN cDNA 2210016F16 gene |
| chr5_-_32746317 | 0.07 |
ENSMUST00000135248.1
|
Pisd
|
phosphatidylserine decarboxylase |
| chr5_-_122989260 | 0.07 |
ENSMUST00000118027.1
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr9_-_110117379 | 0.07 |
ENSMUST00000111991.2
ENSMUST00000149199.1 ENSMUST00000035056.7 ENSMUST00000148287.1 ENSMUST00000127744.1 |
Dhx30
|
DEAH (Asp-Glu-Ala-His) box polypeptide 30 |
| chr4_-_155863362 | 0.07 |
ENSMUST00000030949.3
|
Tas1r3
|
taste receptor, type 1, member 3 |
| chr2_+_85037212 | 0.07 |
ENSMUST00000077798.6
|
Ssrp1
|
structure specific recognition protein 1 |
| chr9_+_110305185 | 0.07 |
ENSMUST00000068071.7
|
Elp6
|
elongator acetyltransferase complex subunit 6 |
| chr12_+_88329254 | 0.06 |
ENSMUST00000091715.1
|
Gm10264
|
predicted gene 10264 |
| chr8_+_45658731 | 0.06 |
ENSMUST00000143820.1
ENSMUST00000132139.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr11_-_94704499 | 0.06 |
ENSMUST00000069852.1
|
Gm11541
|
predicted gene 11541 |
| chr19_+_55316313 | 0.06 |
ENSMUST00000095950.2
|
Vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr4_-_58553311 | 0.06 |
ENSMUST00000107571.1
ENSMUST00000055018.4 |
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr10_+_40883819 | 0.06 |
ENSMUST00000105509.1
|
Wasf1
|
WAS protein family, member 1 |
| chr9_-_107605295 | 0.06 |
ENSMUST00000102529.3
|
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr14_+_45351473 | 0.06 |
ENSMUST00000111835.2
|
Styx
|
serine/threonine/tyrosine interaction protein |
| chr18_-_43373248 | 0.06 |
ENSMUST00000118043.1
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr1_-_125912160 | 0.05 |
ENSMUST00000159417.1
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr2_+_174330006 | 0.05 |
ENSMUST00000109085.1
ENSMUST00000109087.1 ENSMUST00000109084.1 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr11_-_69920892 | 0.05 |
ENSMUST00000152589.1
ENSMUST00000108612.1 ENSMUST00000108611.1 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr11_+_35769462 | 0.05 |
ENSMUST00000018990.7
|
Pank3
|
pantothenate kinase 3 |
| chr12_+_24498570 | 0.05 |
ENSMUST00000075954.7
|
Taf1b
|
TATA box binding protein (Tbp)-associated factor, RNA polymerase I, B |
| chr3_+_88553716 | 0.05 |
ENSMUST00000008748.6
|
Ubqln4
|
ubiquilin 4 |
| chr5_+_91074611 | 0.05 |
ENSMUST00000031324.4
|
Ereg
|
epiregulin |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gli1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.3 | 1.4 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.3 | 0.8 | GO:1904980 | regulation of endosome organization(GO:1904978) positive regulation of endosome organization(GO:1904980) |
| 0.2 | 0.8 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.2 | 0.6 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.2 | 1.4 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 | 0.5 | GO:2000331 | regulation of terminal button organization(GO:2000331) |
| 0.2 | 0.6 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 | 0.8 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 0.1 | 0.6 | GO:0021941 | negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 0.1 | 0.6 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.1 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.6 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.9 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 2.2 | GO:0045605 | negative regulation of epidermal cell differentiation(GO:0045605) |
| 0.1 | 1.7 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.1 | 0.8 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.4 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.1 | 0.5 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.3 | GO:2001074 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.1 | 0.7 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.4 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.1 | 0.3 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.1 | 0.3 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.4 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.6 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.8 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 1.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.5 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.6 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 1.1 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.7 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.2 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.1 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 1.0 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.7 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 1.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.3 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.2 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.6 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.4 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 2.5 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.1 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.6 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.0 | 0.4 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.2 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.6 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.2 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.1 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.1 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.4 | GO:1903077 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.2 | 2.7 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.2 | 0.7 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.2 | 0.6 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.5 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 0.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.3 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 1.3 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.8 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 2.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 0.6 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.2 | 0.7 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.2 | 0.8 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.2 | 0.5 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.2 | 0.6 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 0.8 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 0.6 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 0.8 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 1.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 2.0 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.1 | 0.8 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.6 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 0.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.7 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 1.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.3 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.3 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.0 | 0.1 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 2.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.3 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 2.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 1.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 2.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.8 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 4.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.4 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 1.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.6 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |