Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Glis3
Z-value: 0.58
Transcription factors associated with Glis3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Glis3
|
ENSMUSG00000052942.7 | GLIS family zinc finger 3 |
|
Glis3
|
ENSMUSG00000091294.1 | GLIS family zinc finger 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Glis3 | mm10_v2_chr19_-_28680077_28680122 | -0.32 | 5.6e-02 | Click! |
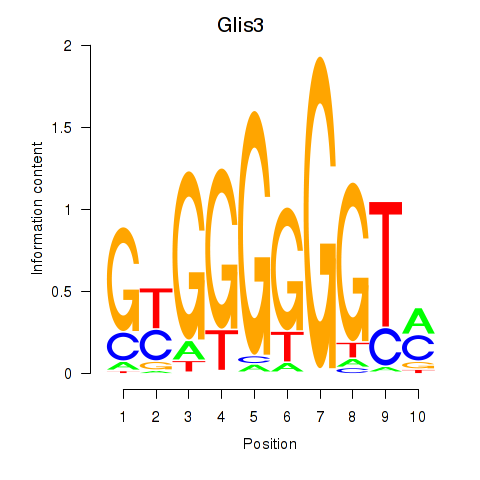
Activity profile of Glis3 motif
Sorted Z-values of Glis3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_55561060 | 2.87 |
ENSMUST00000117701.1
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr14_+_55560904 | 2.66 |
ENSMUST00000072530.4
ENSMUST00000128490.1 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr6_+_17463826 | 2.35 |
ENSMUST00000140070.1
|
Met
|
met proto-oncogene |
| chr14_+_55560480 | 2.18 |
ENSMUST00000121622.1
ENSMUST00000143431.1 ENSMUST00000150481.1 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr4_+_144893077 | 2.16 |
ENSMUST00000154208.1
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr19_+_4711153 | 2.10 |
ENSMUST00000008991.6
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr13_-_29984219 | 2.06 |
ENSMUST00000146092.1
|
E2f3
|
E2F transcription factor 3 |
| chr11_+_101468164 | 1.91 |
ENSMUST00000001347.6
|
Rnd2
|
Rho family GTPase 2 |
| chr4_+_144893127 | 1.90 |
ENSMUST00000142808.1
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr4_+_144892813 | 1.86 |
ENSMUST00000105744.1
ENSMUST00000171001.1 |
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr6_+_17463927 | 1.67 |
ENSMUST00000115442.1
|
Met
|
met proto-oncogene |
| chr6_+_17463749 | 1.62 |
ENSMUST00000115443.1
|
Met
|
met proto-oncogene |
| chr14_-_55560340 | 1.62 |
ENSMUST00000066106.3
|
A730061H03Rik
|
RIKEN cDNA A730061H03 gene |
| chrX_-_136868537 | 1.59 |
ENSMUST00000058814.6
|
Rab9b
|
RAB9B, member RAS oncogene family |
| chr16_+_44173271 | 1.43 |
ENSMUST00000088356.4
ENSMUST00000169582.1 |
Gm608
|
predicted gene 608 |
| chr13_+_119690462 | 1.34 |
ENSMUST00000179869.1
|
Hmgcs1
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| chr2_+_32646586 | 1.30 |
ENSMUST00000009705.7
ENSMUST00000167841.1 |
Eng
|
endoglin |
| chr4_+_86053887 | 1.28 |
ENSMUST00000107178.2
ENSMUST00000048885.5 ENSMUST00000141889.1 ENSMUST00000120678.1 |
Adamtsl1
|
ADAMTS-like 1 |
| chr16_+_20733104 | 1.27 |
ENSMUST00000115423.1
ENSMUST00000007171.6 |
Chrd
|
chordin |
| chr6_-_28134545 | 1.25 |
ENSMUST00000115323.1
|
Grm8
|
glutamate receptor, metabotropic 8 |
| chr10_-_68278713 | 1.24 |
ENSMUST00000020106.7
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr2_-_29253001 | 1.20 |
ENSMUST00000071201.4
|
Ntng2
|
netrin G2 |
| chr13_+_38345716 | 1.05 |
ENSMUST00000171970.1
|
Bmp6
|
bone morphogenetic protein 6 |
| chr4_+_118428078 | 1.03 |
ENSMUST00000006557.6
ENSMUST00000167636.1 ENSMUST00000102673.4 |
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chr1_-_168431695 | 0.99 |
ENSMUST00000176790.1
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr15_-_39857459 | 0.93 |
ENSMUST00000022915.3
ENSMUST00000110306.1 |
Dpys
|
dihydropyrimidinase |
| chr6_-_122820606 | 0.93 |
ENSMUST00000181317.1
|
Gm26826
|
predicted gene, 26826 |
| chrX_+_71364901 | 0.90 |
ENSMUST00000132837.1
|
Mtmr1
|
myotubularin related protein 1 |
| chr1_-_168432270 | 0.89 |
ENSMUST00000072863.4
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr11_-_102296618 | 0.88 |
ENSMUST00000107132.2
ENSMUST00000073234.2 |
Atxn7l3
|
ataxin 7-like 3 |
| chr5_-_72974702 | 0.85 |
ENSMUST00000043711.8
|
Gm10135
|
predicted gene 10135 |
| chr2_+_131234043 | 0.84 |
ENSMUST00000041362.5
ENSMUST00000110199.2 |
Mavs
|
mitochondrial antiviral signaling protein |
| chr5_-_91402905 | 0.82 |
ENSMUST00000121044.2
|
Btc
|
betacellulin, epidermal growth factor family member |
| chr9_+_31280525 | 0.81 |
ENSMUST00000117389.1
|
Prdm10
|
PR domain containing 10 |
| chr3_+_66981352 | 0.81 |
ENSMUST00000162036.1
|
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr10_+_93641041 | 0.80 |
ENSMUST00000020204.4
|
Ntn4
|
netrin 4 |
| chr15_+_102406143 | 0.79 |
ENSMUST00000170884.1
ENSMUST00000165924.1 ENSMUST00000163709.1 ENSMUST00000001326.6 |
Sp1
|
trans-acting transcription factor 1 |
| chrX_+_71364745 | 0.79 |
ENSMUST00000114601.1
ENSMUST00000146213.1 ENSMUST00000015358.1 |
Mtmr1
|
myotubularin related protein 1 |
| chr4_-_41695935 | 0.77 |
ENSMUST00000145379.1
|
Cntfr
|
ciliary neurotrophic factor receptor |
| chrM_+_9452 | 0.76 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr11_-_97187872 | 0.74 |
ENSMUST00000001479.4
|
Kpnb1
|
karyopherin (importin) beta 1 |
| chr11_+_98927785 | 0.70 |
ENSMUST00000107474.1
|
Rara
|
retinoic acid receptor, alpha |
| chr9_-_29412204 | 0.68 |
ENSMUST00000115237.1
|
Ntm
|
neurotrimin |
| chr5_+_104459450 | 0.64 |
ENSMUST00000086831.3
|
Pkd2
|
polycystic kidney disease 2 |
| chr11_-_72489904 | 0.64 |
ENSMUST00000045303.3
|
Spns2
|
spinster homolog 2 |
| chr11_+_97663366 | 0.60 |
ENSMUST00000044730.5
|
Mllt6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chrX_-_38564519 | 0.59 |
ENSMUST00000016681.8
|
Cul4b
|
cullin 4B |
| chr15_+_102921103 | 0.59 |
ENSMUST00000001700.6
|
Hoxc13
|
homeobox C13 |
| chr7_-_49636847 | 0.57 |
ENSMUST00000032717.6
|
Dbx1
|
developing brain homeobox 1 |
| chr17_+_56326045 | 0.56 |
ENSMUST00000139679.1
ENSMUST00000025036.4 ENSMUST00000086835.5 |
Kdm4b
|
lysine (K)-specific demethylase 4B |
| chr17_-_70851189 | 0.56 |
ENSMUST00000059775.8
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr1_-_64121389 | 0.55 |
ENSMUST00000055001.3
|
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr13_+_119428583 | 0.55 |
ENSMUST00000109203.2
|
Paip1
|
polyadenylate binding protein-interacting protein 1 |
| chr9_-_48835932 | 0.53 |
ENSMUST00000093852.3
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr14_+_25607797 | 0.53 |
ENSMUST00000160229.1
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr15_+_102407144 | 0.53 |
ENSMUST00000169619.1
|
Sp1
|
trans-acting transcription factor 1 |
| chr7_+_46796088 | 0.53 |
ENSMUST00000006774.4
ENSMUST00000165031.1 |
Gtf2h1
|
general transcription factor II H, polypeptide 1 |
| chr2_-_73892588 | 0.51 |
ENSMUST00000154456.1
ENSMUST00000090802.4 ENSMUST00000055833.5 |
Atf2
|
activating transcription factor 2 |
| chr6_-_83572429 | 0.51 |
ENSMUST00000068054.7
|
Stambp
|
STAM binding protein |
| chr5_+_110135823 | 0.50 |
ENSMUST00000112519.2
ENSMUST00000014812.8 |
Chfr
|
checkpoint with forkhead and ring finger domains |
| chr3_+_52268337 | 0.50 |
ENSMUST00000053764.5
|
Foxo1
|
forkhead box O1 |
| chr11_+_98795495 | 0.50 |
ENSMUST00000037915.2
|
Msl1
|
male-specific lethal 1 homolog (Drosophila) |
| chr17_-_34031644 | 0.48 |
ENSMUST00000171872.1
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr9_-_108305941 | 0.46 |
ENSMUST00000044725.7
|
Tcta
|
T cell leukemia translocation altered gene |
| chr17_-_34031544 | 0.44 |
ENSMUST00000025186.8
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr2_-_73892619 | 0.44 |
ENSMUST00000112007.1
ENSMUST00000112016.2 |
Atf2
|
activating transcription factor 2 |
| chr6_+_108660616 | 0.43 |
ENSMUST00000032194.4
|
Bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr10_+_61175206 | 0.43 |
ENSMUST00000079235.5
|
Tbata
|
thymus, brain and testes associated |
| chr13_+_37826018 | 0.42 |
ENSMUST00000110238.2
|
Rreb1
|
ras responsive element binding protein 1 |
| chr1_-_93445642 | 0.41 |
ENSMUST00000042498.7
|
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr13_+_37825975 | 0.40 |
ENSMUST00000138043.1
|
Rreb1
|
ras responsive element binding protein 1 |
| chr17_+_80944611 | 0.40 |
ENSMUST00000025092.4
|
Tmem178
|
transmembrane protein 178 |
| chr17_-_34031684 | 0.39 |
ENSMUST00000169397.1
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chrX_-_101419788 | 0.39 |
ENSMUST00000117901.1
ENSMUST00000120201.1 ENSMUST00000117637.1 ENSMUST00000134005.1 ENSMUST00000121520.1 |
Zmym3
|
zinc finger, MYM-type 3 |
| chr6_+_108660772 | 0.39 |
ENSMUST00000163617.1
|
Bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr16_+_20673264 | 0.35 |
ENSMUST00000154950.1
ENSMUST00000115461.1 |
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr3_+_121426495 | 0.34 |
ENSMUST00000029773.8
|
Cnn3
|
calponin 3, acidic |
| chr1_-_168431502 | 0.34 |
ENSMUST00000064438.4
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr11_-_69801716 | 0.34 |
ENSMUST00000011285.4
ENSMUST00000102585.1 |
Fgf11
|
fibroblast growth factor 11 |
| chr16_+_20673517 | 0.33 |
ENSMUST00000115460.1
|
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr7_+_90442729 | 0.33 |
ENSMUST00000061767.4
ENSMUST00000107206.1 |
Crebzf
|
CREB/ATF bZIP transcription factor |
| chr3_+_89245952 | 0.31 |
ENSMUST00000040888.5
|
Krtcap2
|
keratinocyte associated protein 2 |
| chr4_+_137993445 | 0.30 |
ENSMUST00000105831.2
ENSMUST00000084214.5 |
Eif4g3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr11_-_70237761 | 0.29 |
ENSMUST00000108576.3
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr11_+_116030304 | 0.29 |
ENSMUST00000021116.5
ENSMUST00000106452.1 |
Unk
|
unkempt homolog (Drosophila) |
| chrX_+_151522352 | 0.27 |
ENSMUST00000148622.1
|
Phf8
|
PHD finger protein 8 |
| chr11_-_85139939 | 0.26 |
ENSMUST00000108075.2
|
Usp32
|
ubiquitin specific peptidase 32 |
| chr17_+_86963279 | 0.26 |
ENSMUST00000139344.1
|
Rhoq
|
ras homolog gene family, member Q |
| chr2_-_73892530 | 0.26 |
ENSMUST00000136958.1
ENSMUST00000112010.2 ENSMUST00000128531.1 ENSMUST00000112017.1 |
Atf2
|
activating transcription factor 2 |
| chr5_-_24527276 | 0.26 |
ENSMUST00000088311.4
|
Gbx1
|
gastrulation brain homeobox 1 |
| chr7_+_125368861 | 0.24 |
ENSMUST00000166958.1
|
4930571K23Rik
|
RIKEN cDNA 4930571K23 gene |
| chr3_-_94582716 | 0.23 |
ENSMUST00000029783.9
|
Snx27
|
sorting nexin family member 27 |
| chr3_-_142881942 | 0.23 |
ENSMUST00000043812.8
|
Pkn2
|
protein kinase N2 |
| chr2_-_144331695 | 0.21 |
ENSMUST00000103171.3
|
Ovol2
|
ovo-like 2 (Drosophila) |
| chr17_+_37045963 | 0.21 |
ENSMUST00000025338.9
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr7_-_46795881 | 0.21 |
ENSMUST00000107653.1
ENSMUST00000107654.1 ENSMUST00000014562.7 ENSMUST00000152759.1 |
Hps5
|
Hermansky-Pudlak syndrome 5 homolog (human) |
| chrM_+_8600 | 0.20 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr10_+_127739516 | 0.20 |
ENSMUST00000054287.7
|
Zbtb39
|
zinc finger and BTB domain containing 39 |
| chr3_-_94582548 | 0.20 |
ENSMUST00000107283.1
|
Snx27
|
sorting nexin family member 27 |
| chr8_-_87472576 | 0.19 |
ENSMUST00000034076.8
|
Cbln1
|
cerebellin 1 precursor protein |
| chr3_+_89246397 | 0.19 |
ENSMUST00000168900.1
|
Krtcap2
|
keratinocyte associated protein 2 |
| chr13_+_119428888 | 0.18 |
ENSMUST00000026520.7
|
Paip1
|
polyadenylate binding protein-interacting protein 1 |
| chr4_-_120747248 | 0.18 |
ENSMUST00000030376.7
|
Kcnq4
|
potassium voltage-gated channel, subfamily Q, member 4 |
| chr9_+_107928440 | 0.16 |
ENSMUST00000085073.1
|
Actl11
|
actin-like 11 |
| chr11_-_70237852 | 0.15 |
ENSMUST00000108575.2
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr11_-_70237886 | 0.15 |
ENSMUST00000108577.1
ENSMUST00000108579.1 ENSMUST00000021181.6 ENSMUST00000108578.2 ENSMUST00000102569.3 |
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr13_-_40730416 | 0.15 |
ENSMUST00000021787.5
|
Tfap2a
|
transcription factor AP-2, alpha |
| chr17_-_23745829 | 0.15 |
ENSMUST00000046525.8
|
Kremen2
|
kringle containing transmembrane protein 2 |
| chr11_-_97744659 | 0.15 |
ENSMUST00000018691.8
|
Pip4k2b
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta |
| chr9_+_108306205 | 0.14 |
ENSMUST00000007959.8
|
Rhoa
|
ras homolog gene family, member A |
| chr7_-_126200413 | 0.14 |
ENSMUST00000163959.1
|
Xpo6
|
exportin 6 |
| chr4_+_129058133 | 0.13 |
ENSMUST00000030584.4
ENSMUST00000168461.1 ENSMUST00000152565.1 |
Rnf19b
|
ring finger protein 19B |
| chrX_-_23365044 | 0.12 |
ENSMUST00000115313.1
|
Klhl13
|
kelch-like 13 |
| chr10_+_63457505 | 0.12 |
ENSMUST00000105440.1
|
Ctnna3
|
catenin (cadherin associated protein), alpha 3 |
| chr11_+_69632927 | 0.12 |
ENSMUST00000018909.3
|
Fxr2
|
fragile X mental retardation, autosomal homolog 2 |
| chr11_-_52000432 | 0.12 |
ENSMUST00000020657.6
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chrX_-_111697069 | 0.11 |
ENSMUST00000113422.2
ENSMUST00000038472.5 |
Hdx
|
highly divergent homeobox |
| chrX_+_105764262 | 0.10 |
ENSMUST00000033581.3
|
Fgf16
|
fibroblast growth factor 16 |
| chr13_+_37826225 | 0.09 |
ENSMUST00000128570.1
|
Rreb1
|
ras responsive element binding protein 1 |
| chr19_-_6987621 | 0.09 |
ENSMUST00000130048.1
ENSMUST00000025914.6 |
Vegfb
|
vascular endothelial growth factor B |
| chrX_-_135009185 | 0.08 |
ENSMUST00000113185.2
ENSMUST00000064659.5 |
Zmat1
|
zinc finger, matrin type 1 |
| chr3_+_96601084 | 0.08 |
ENSMUST00000062058.3
|
Lix1l
|
Lix1-like |
| chr8_+_70863127 | 0.07 |
ENSMUST00000050921.2
|
A230052G05Rik
|
RIKEN cDNA A230052G05 gene |
| chr9_-_110117379 | 0.06 |
ENSMUST00000111991.2
ENSMUST00000149199.1 ENSMUST00000035056.7 ENSMUST00000148287.1 ENSMUST00000127744.1 |
Dhx30
|
DEAH (Asp-Glu-Ala-His) box polypeptide 30 |
| chr17_-_65613521 | 0.06 |
ENSMUST00000024897.8
|
Vapa
|
vesicle-associated membrane protein, associated protein A |
| chr11_-_70237638 | 0.04 |
ENSMUST00000100950.3
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr2_-_119477613 | 0.04 |
ENSMUST00000110808.1
ENSMUST00000049920.7 |
Ino80
|
INO80 homolog (S. cerevisiae) |
| chr6_+_145934113 | 0.04 |
ENSMUST00000032383.7
|
Sspn
|
sarcospan |
| chr8_-_88362154 | 0.04 |
ENSMUST00000034085.7
|
Brd7
|
bromodomain containing 7 |
| chr7_-_46795661 | 0.04 |
ENSMUST00000123725.1
|
Hps5
|
Hermansky-Pudlak syndrome 5 homolog (human) |
| chr7_+_25268387 | 0.04 |
ENSMUST00000169392.1
|
Cic
|
capicua homolog (Drosophila) |
| chr19_+_46305682 | 0.03 |
ENSMUST00000111881.2
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr12_+_102949450 | 0.03 |
ENSMUST00000179002.1
|
Unc79
|
unc-79 homolog (C. elegans) |
| chr11_-_52000748 | 0.03 |
ENSMUST00000109086.1
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr7_+_45017953 | 0.03 |
ENSMUST00000044111.7
|
Rras
|
Harvey rat sarcoma oncogene, subgroup R |
| chr2_-_144332146 | 0.03 |
ENSMUST00000037423.3
|
Ovol2
|
ovo-like 2 (Drosophila) |
| chr11_+_77216180 | 0.03 |
ENSMUST00000037912.5
ENSMUST00000156488.1 |
Ssh2
|
slingshot homolog 2 (Drosophila) |
| chr11_+_96271453 | 0.03 |
ENSMUST00000000010.8
ENSMUST00000174042.1 |
Hoxb9
|
homeobox B9 |
| chr12_+_69241832 | 0.02 |
ENSMUST00000063445.6
|
Klhdc1
|
kelch domain containing 1 |
| chr4_+_63215402 | 0.02 |
ENSMUST00000036300.6
|
Col27a1
|
collagen, type XXVII, alpha 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Glis3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.6 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.4 | 1.3 | GO:1905072 | detection of oxygen(GO:0003032) cardiac jelly development(GO:1905072) |
| 0.4 | 6.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.3 | 0.9 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.3 | 0.8 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.3 | 1.3 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.2 | 1.2 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.2 | 0.7 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.2 | 0.6 | GO:0003127 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) cellular response to water stimulus(GO:0071462) |
| 0.2 | 0.6 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.2 | 0.8 | GO:0003360 | brainstem development(GO:0003360) |
| 0.2 | 1.3 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.2 | 0.7 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.2 | 1.0 | GO:0032349 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.2 | 0.8 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.2 | 1.4 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 1.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.9 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 1.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 1.9 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.1 | GO:0070342 | brown fat cell proliferation(GO:0070342) regulation of brown fat cell proliferation(GO:0070347) |
| 0.1 | 0.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) eye pigmentation(GO:0048069) |
| 0.1 | 0.3 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.1 | 1.0 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.5 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.1 | 0.5 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.1 | GO:0043366 | beta selection(GO:0043366) |
| 0.1 | 0.5 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) positive regulation of NK T cell differentiation(GO:0051138) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.3 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 1.9 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 0.6 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.1 | 0.8 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.1 | 1.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.7 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.4 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.1 | 1.3 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.2 | GO:0021622 | oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.1 | 1.4 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 0.6 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.6 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 1.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.5 | GO:0055093 | response to hyperoxia(GO:0055093) cellular response to hyperoxia(GO:0071455) |
| 0.0 | 0.2 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.9 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.6 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.2 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.2 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.0 | 0.5 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.5 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.8 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.0 | 0.5 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 0.5 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 1.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.8 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.4 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.2 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.0 | 0.8 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.5 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.0 | GO:0002268 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 8.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 0.8 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 0.6 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.5 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 5.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.2 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.5 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.6 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 1.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 1.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 3.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.8 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.0 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.8 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.6 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.8 | 5.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 1.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 1.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 1.3 | GO:0005534 | galactose binding(GO:0005534) |
| 0.2 | 1.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.2 | 0.6 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 0.6 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.2 | 1.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.9 | GO:0016812 | nucleobase binding(GO:0002054) hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 1.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.8 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.1 | 0.8 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.8 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 1.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 1.0 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.7 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.5 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 1.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 1.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.3 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.8 | GO:0047485 | protein N-terminus binding(GO:0047485) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 7.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 2.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 2.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 5.8 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 1.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 1.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.7 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 2.1 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |