Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
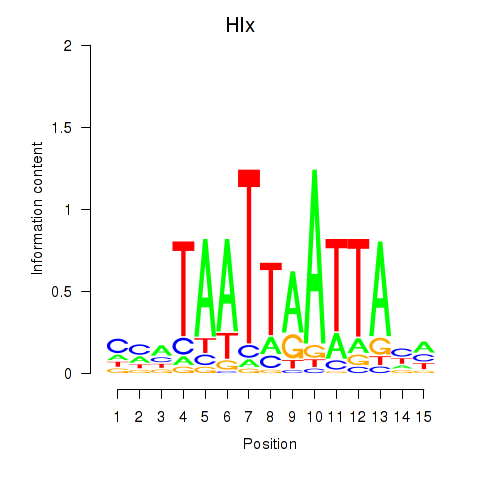
Results for Hlx
Z-value: 0.24
Transcription factors associated with Hlx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hlx
|
ENSMUSG00000039377.6 | H2.0-like homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hlx | mm10_v2_chr1_-_184732616_184732627 | -0.34 | 4.4e-02 | Click! |
Activity profile of Hlx motif
Sorted Z-values of Hlx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_145032765 | 1.09 |
ENSMUST00000029919.5
|
Clca3
|
chloride channel calcium activated 3 |
| chrX_-_143933089 | 0.75 |
ENSMUST00000087313.3
|
Dcx
|
doublecortin |
| chrX_-_143933204 | 0.40 |
ENSMUST00000112851.1
ENSMUST00000112856.2 ENSMUST00000033642.3 |
Dcx
|
doublecortin |
| chr6_+_7555053 | 0.36 |
ENSMUST00000090679.2
ENSMUST00000184986.1 |
Tac1
|
tachykinin 1 |
| chr7_-_45103747 | 0.23 |
ENSMUST00000003512.7
|
Fcgrt
|
Fc receptor, IgG, alpha chain transporter |
| chr11_+_99873389 | 0.22 |
ENSMUST00000093936.3
|
Krtap9-1
|
keratin associated protein 9-1 |
| chr9_+_54980880 | 0.20 |
ENSMUST00000093844.3
|
Chrna5
|
cholinergic receptor, nicotinic, alpha polypeptide 5 |
| chr10_-_107486077 | 0.17 |
ENSMUST00000000445.1
|
Myf5
|
myogenic factor 5 |
| chr9_+_7571396 | 0.16 |
ENSMUST00000120900.1
ENSMUST00000093896.3 ENSMUST00000151853.1 ENSMUST00000152878.1 |
Mmp27
|
matrix metallopeptidase 27 |
| chr5_-_73632421 | 0.12 |
ENSMUST00000087177.2
|
Lrrc66
|
leucine rich repeat containing 66 |
| chr19_+_53944870 | 0.12 |
ENSMUST00000025932.7
|
Shoc2
|
soc-2 (suppressor of clear) homolog (C. elegans) |
| chrM_+_2743 | 0.10 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr15_-_8710734 | 0.09 |
ENSMUST00000005493.7
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr10_+_102158858 | 0.09 |
ENSMUST00000138522.1
ENSMUST00000163753.1 ENSMUST00000138016.1 |
Mgat4c
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr10_+_102159000 | 0.08 |
ENSMUST00000020039.6
|
Mgat4c
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr3_+_32436376 | 0.07 |
ENSMUST00000108242.1
|
Pik3ca
|
phosphatidylinositol 3-kinase, catalytic, alpha polypeptide |
| chr11_-_99244058 | 0.07 |
ENSMUST00000103132.3
ENSMUST00000038214.6 |
Krt222
|
keratin 222 |
| chr2_+_78051155 | 0.06 |
ENSMUST00000145972.1
|
4930440I19Rik
|
RIKEN cDNA 4930440I19 gene |
| chr19_-_53944621 | 0.06 |
ENSMUST00000135402.2
|
Bbip1
|
BBSome interacting protein 1 |
| chr7_-_101837776 | 0.06 |
ENSMUST00000165052.1
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr10_-_37138863 | 0.05 |
ENSMUST00000092584.5
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr1_+_180109192 | 0.05 |
ENSMUST00000143176.1
ENSMUST00000135056.1 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr5_-_62766153 | 0.04 |
ENSMUST00000076623.4
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chrX_+_133850980 | 0.04 |
ENSMUST00000033602.8
|
Tnmd
|
tenomodulin |
| chr5_+_121749196 | 0.02 |
ENSMUST00000161064.1
|
Atxn2
|
ataxin 2 |
| chr15_-_8710409 | 0.00 |
ENSMUST00000157065.1
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hlx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2000851 | positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.1 | 1.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |